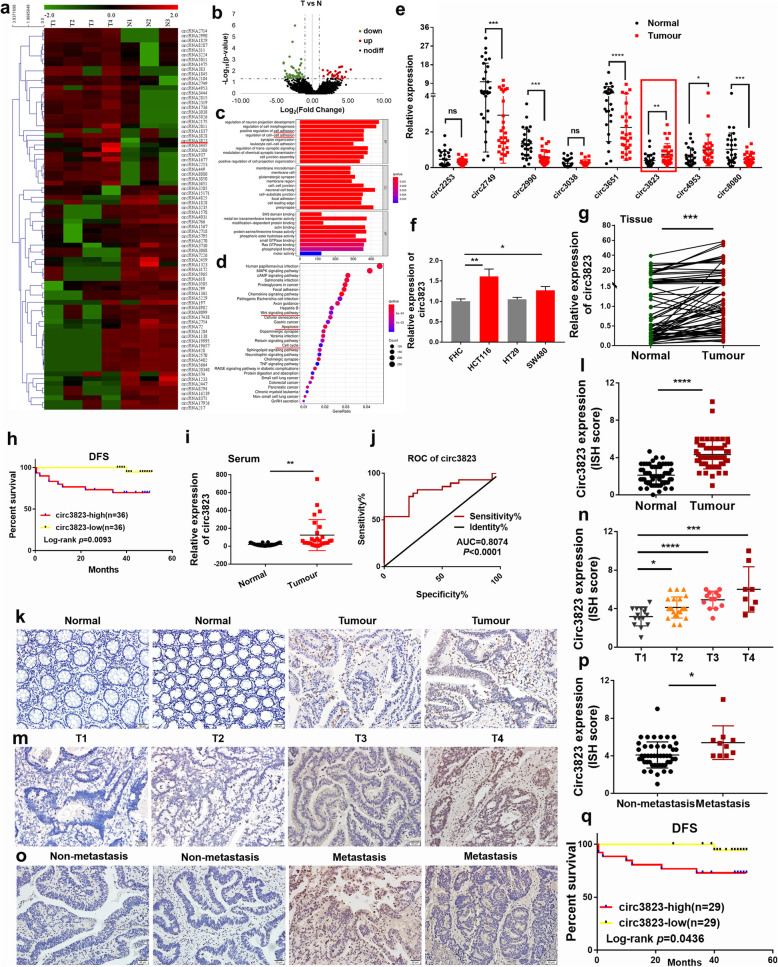
Fig. 1.
CircRNA expression profiling in CRC tissues and normal tissues and validation of the circRNAs expression in serums and tissues (T: tumour, N: normal). a Hierarchical clustering of differentially expressed circRNA in two groups. b Volcano plot showed the expression profile between tumour tissue and normal tissue. Conditions for screening differences:│Fold Change│ > 2; p < 0.05. The red points in the plot indicated significantly up-regulated circRNAs, and the green points indicated significantly down-regulated circRNAs. c GO function analysis of differentially expressed circRNAs. d KEGG pathway analysis of differentially expressed circRNAs. e The expression of circ2253, circ2749, circ2990, circ3038, circ3651, circ3823, circ4953 and circ8080 was detected by qRT-PCR in 28 CRC tissues and matched normal tissues. f The expression of circ3823 was measured by qRT-PCR in CRC cells and FHC. g The level of circ3823 in 72 CRC tissues and matched normal tissues were analyzed by qRT-PCR. h The disease-free survival analysis of 72 CRC patients was performed based on qRT-PCR data. Log-rank test was used to estimate the significance. i The expression of circ3823 was measured by qRT-PCR in 28 serum samples from CRC patients and 28 serum samples from healthy people. GAPDH was used as an internal reference. j ROC curves based on the expression of circ3823 in serum. k, l ISH images and ISH scores statistical analysis of circ3823 in CRC tissues and normal tissues. m, n ISH images and ISH scores statistical analysis of circ3823 in T1, T2, T3, T4 CRC tissues. o, p ISH images and ISH scores statistical analysis of circ3823 in lymph node metastasis tissues and non-lymph node metastasis tissues (magnification, × 200, scale bar, 50 μm). q The disease-free survival analysis of 58 CRC patients based on circ3823 ISH scores. Log-rank test was used to estimate the significance. Data were indicated as mean ± SD, ns P ≥ 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001