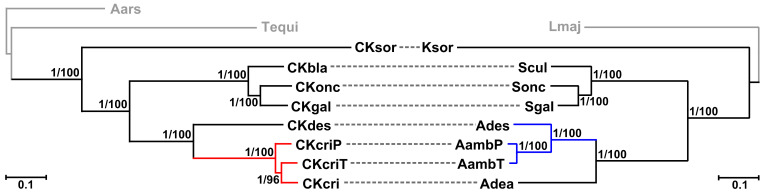
Figure 3.
Juxtaposed maximum-likelihood phylogenomic trees of endosymbiotic bacteria and their respective trypanosomatid hosts. Outgroups are shown in grey. Dashed lines connect endosymbionts with their hosts, while colored branches point to the discrepancy between their phylogenies. Numbers at branches indicate bootstrap support and Bayesian posterior probability values, respectively. Scale bars show the number of substitutions per site. Organism codes: Lmaj, Leishmania major; Ksor, Kentomonas sorsogonicus; Scul, Strigomonas culicis; Sonc, S. oncopelti; Sgal, S. galati; Ades, Angomonas desouzai; AambP and AambT, A. ambiguus strains PNG-M02 and TCC2535, respectively; Adea, A. deanei; Aars, Achromobacter arsenitoxydans; Tequi, Taylorella equigenitalis; CKsor, Candidatus Kinetoplastibacterium sorsogonicusi; CKbla, Ca. K. blastocrithidii; CKonc, Ca. K. oncopeltii; CKgal, Ca. K. galatii; CKdes, Ca. K. desouzaii; CKcri, CKcriP, and CKcriT, Ca. K crithidii TCC036E, PNG-M02, and TCC2435, respectively.