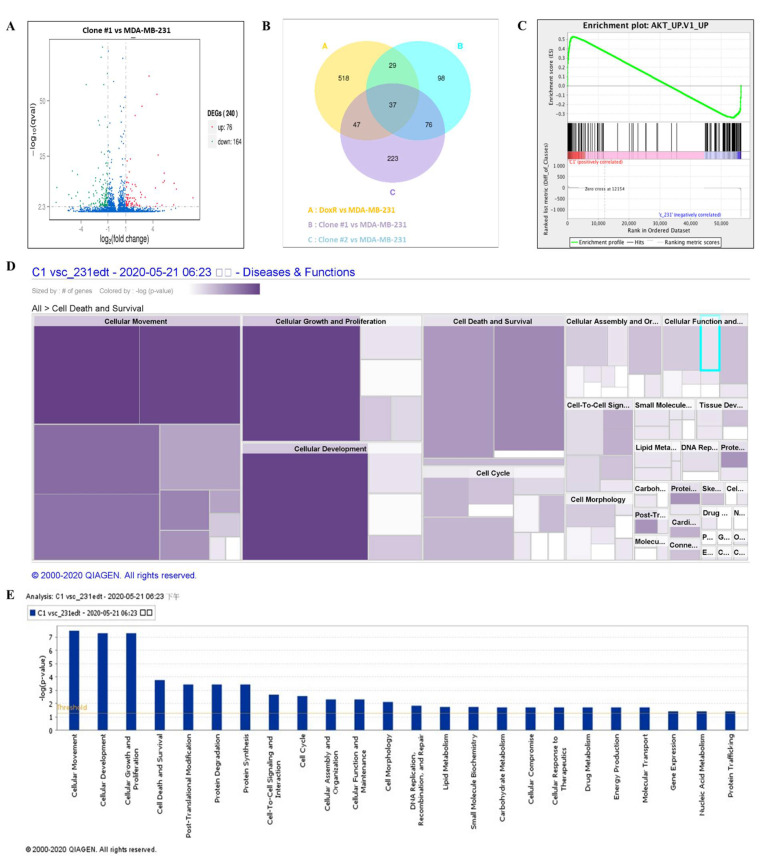
Figure 7.
Differential gene expression analysis and the potential mechanism of DEHP-induced drug resistance. (A) Volcano plot of the differentially expressed genes between DEHP-exposed clone #1 and MDA-MB-231 cells plotted by log2 (fold change) with q value < 0.05. Significantly differentially expressed genes are represented by ⬤ red dots (upregulation) and ⬤ green dots (downregulation). The x-axis represents the genes expressed in the samples, and the y-axis is the significance of the degree of expression. (B) The Venn Diagram of Differential Gene Expression, it presents the number of differentially expressed genes in each group and the overlapping genes between different groups. Group A: DoxR vs. MDA-MB-231 (yellow), Group B: Clone #1 vs. MDA-MB-231 (blue), and Group C: Clone #2 vs. MDA-MB-231 (violet). (C) Gene set enrichment analysis showing enrichment plot of Akt signaling with positive correlation of clone #1. Ingenuity pathway analysis (IPA-Qiagen) of the NGS data to compare the disease and molecular function of DEHP-exposed clone #1 with those of parental MDA-MB-231 cells. (D) Heatmap analysis of the top 20 cellular diseases and functions of the DEGs involved. Size represents the number of DEGs, and color by –log (p-value) represents the associated log of the calculated p-value. The color intensity (purple color) of the heatmap squares is proportional to the significance of the p-value. A smaller p-value (means a larger –log of that value) indicates a more significant association of genes. The p-value was calculated using right-tailed Fisher’s exact test. (E) Bar graph analysis of the top 20 cellular and molecular functions of the DEGs involved. The X-axis represents cellular or molecular functions, and the Y-axis represents –log (p-value). The yellow line represents the threshold.