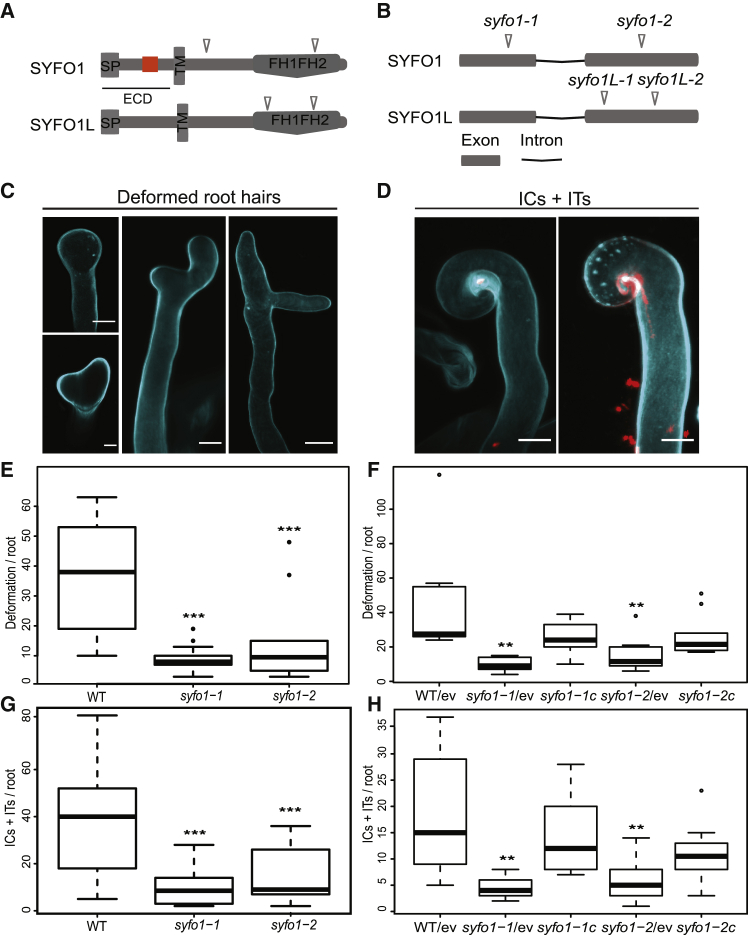
Figure 1.
syfo1 mutants are impaired in symbiotic root hair responses
(A and B) Isolation of independent mutant alleles with Tnt1 transposon insertions mapping to different regions of the SYFO1 and SYFO1L protein (A) and the gene (B) models. ECD, extracellular domain; SP, signal peptide; TM, transmembrane domain; orange box, proline-rich repeat (PRR).
(C and D) Images show cell wall stained by Calcofluor white in deformed root hairs (C), infection chambers (ICs), and infection threads (ITs) (D) on wild-type plants to illustrate the scored structures. Scale bars indicate 10 μm.
(E and F) syfo1 mutants show significantly reduced responsiveness to the presence of compatible rhizobia when assessing root hair deformations in mutants (E) and genetically complemented roots by introducing a genomic version of the full-length SYFO1 gene driven by the endogenous SYFO1 promoter (syfo1-1c and syfo1-2c) with an empty vector (ev) transformation control aside (F).
(G and H) Infection-related structures, such as ICs + ITs, were scored in mutants (G) and complemented roots (H).
Asterisks indicate a significant statistical difference based on a Tukey-Kramer multiple-comparison test with ∗∗p < 0.01 and ∗∗∗p < 0.001. Data are shown as mean ± SE with independent 9–14 plants for phenotypical analysis and 10 plants for complementation analysis. Phenotypes were scored at 5 dpi. See also Figures S1–S3 and Table S1.