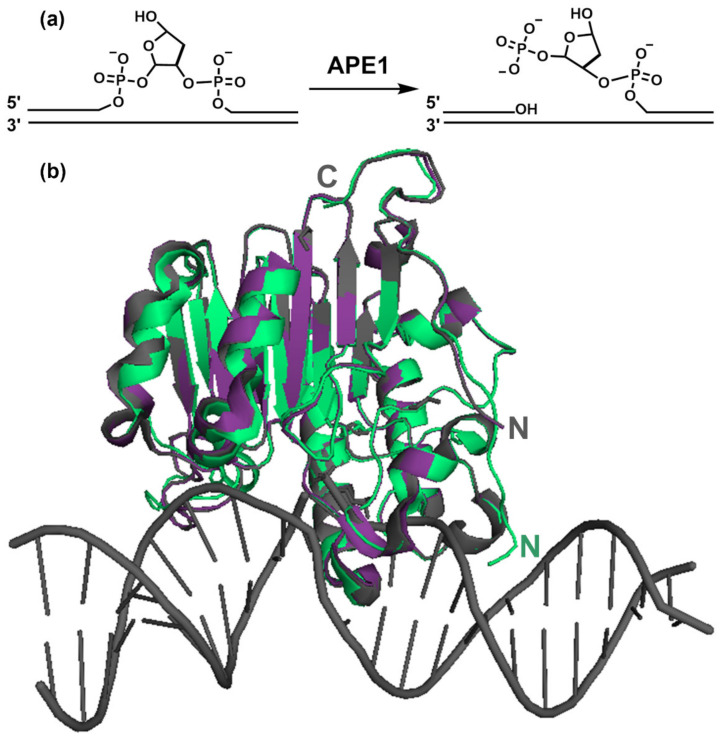
Figure 2.
(a) Incision reaction catalyzed by APE1; (b) APE1 3D structure. Structural models of human APE1 either free, in green (accession code 4QHE) [49], or bound to abasic substrate and product-mimicking DNA oligonucleotides (magenta, 5DFI and grey, 5DFF, respectively [16]). The DNA structure corresponds to entry 5DFF. The structures of the substrate and product complexes have a root mean square deviation (r.m.s.d.) of 0.62 Å between them (referred to Cα atoms) and 1.19 Å and 1.36 Å as compared with free APE1, respectively. “N” and “C” indicate the N- and C- termini, respectively. The models start with residue 38 (4QHE) or 43 (5DFI and 5DFF), and thus do not provide information on the N-terminal tail of APE1. The figure was prepared with PyMol [50].