Figure 5. End processing can occur on double-stranded DNA with mismatched 3′-termini.
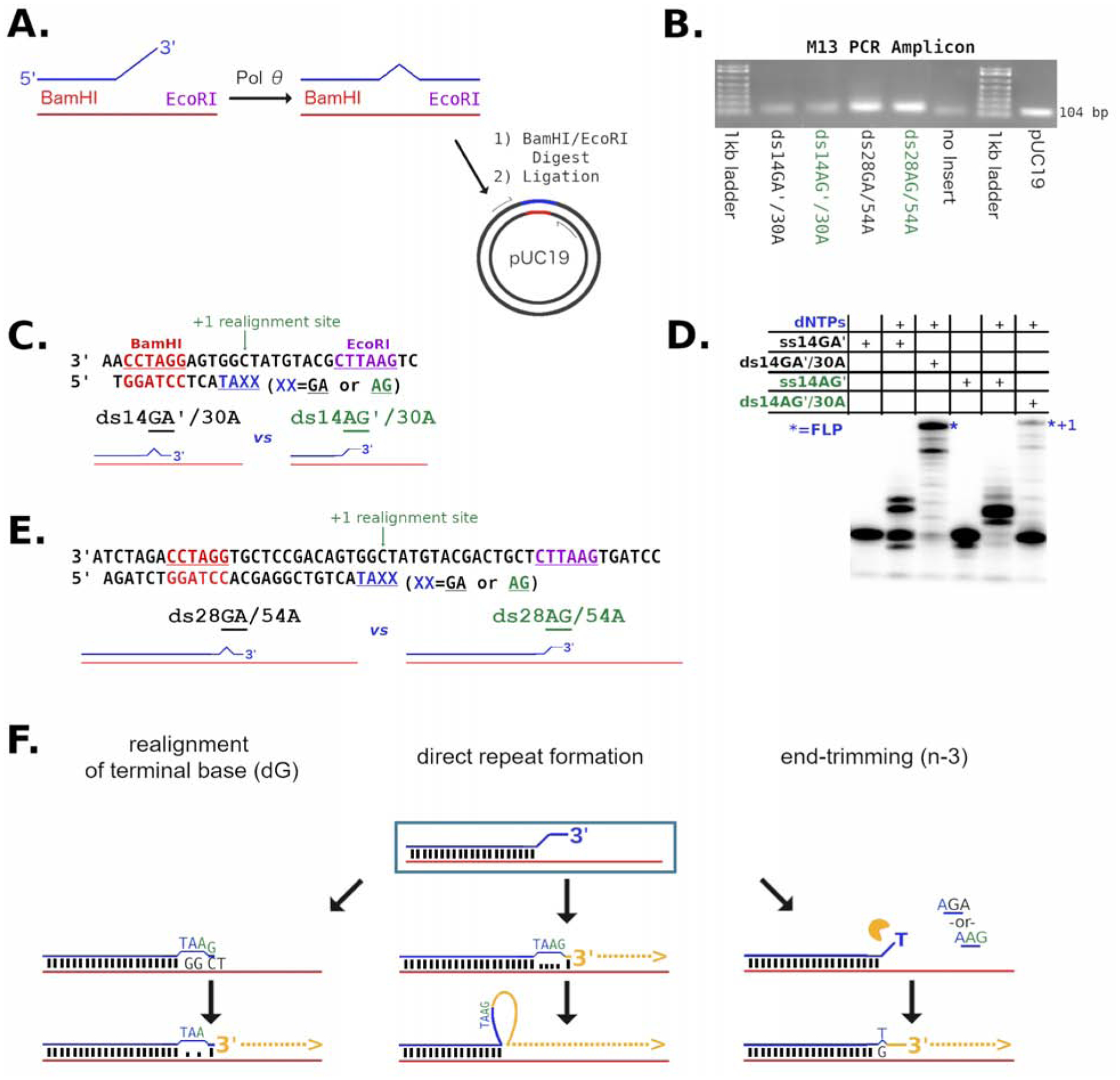
A) Strategy for pol QM1 reaction with substrates in vitro prior to sequencing analysis. The primer (blue) is annealed to a template (red) with a BamHI (also red) recognition site in the duplex region. Full extension of the primer by pol θ forms an EcoRI (purple) sequence. Following dual restriction digestion, products are ligated into pUC19. Various products may be formed (illustrated by the bulge), arising from mismatches left or editing performed by pol θ. B) PCR with m13 primers generates amplicons of approximately 100 bp which are then subjected to DNA sequencing. C) The substrates ds14′GA/30A (black) and ds14′AG/30A (green) are diagrammed D) Reactions of ds14GA/30A and ds14AG′/30A are compared on a denaturing gel as indicated. The FLP (*) marked by “n+1” is a single nucleotide longer with the ds14AG/30A substrate. E) The 28/54 series of substrates are described graphically to show that the only difference between ds28GA/54A (black) and ds28AG/54A (green) is the two 3′ bases of the primer at the 5′-overhang. F) The predominant ways by which pol θ processes the substrates are realignment of the terminus (left), direct repeat formation (center), and end-trimming (right). See also Figure S4.
