Abstract
Background
Postsurgical infections due to Mycobacterium chimaera appeared as a novel nosocomial threat in 2015, with a worldwide outbreak due to contaminated heater-cooler units used in open chest surgery. We report the results of investigations conducted in France including whole-genome sequencing comparison of patient and heater-cooler unit isolates.
Methods
We sought M. chimaera infection cases from 2010 onwards through national epidemiological investigations in health care facilities performing cardiopulmonary bypass, together with a survey on good practices and systematic heater-cooler unit microbial analyses. Clinical and heater-cooler unit isolates were subjected to whole-genome sequencing analyzed with regard to the reference outbreak strain Zuerich-1.
Results
Only 2 clinical cases were shown to be related to the outbreak, although 23% (41/175) of heater-cooler units were declared positive for M. avium complex. Specific measures to prevent infection were applied in 89% (50/56) of health care facilities, although only 14% (8/56) of them followed the manufacturer maintenance recommendations. Whole-genome sequencing comparison showed that the clinical isolates and 72% (26/36) of heater-cooler unit isolates belonged to the epidemic cluster. Within clinical isolates, 5–9 nonsynonymous single nucleotide polymorphisms were observed, among which an in vivo mutation in a putative efflux pump gene was observed in a clinical isolate obtained for 1 patient on antimicrobial treatment.
Conclusions
Cases of postsurgical M. chimaera infections have been declared to be rare in France, although heater-cooler units were contaminated, as in other countries. Genomic analyses confirmed the connection to the outbreak and identified specific single nucleotide polymorphisms, including 1 suggesting fitness evolution in vivo.
Keywords: heater-cooler units (HCUs), mmpL, nontuberculous mycobacteria (NTM), molecular epidemiology
Since the Rapid Risk Assessment (RRA) alert issued by the European Centre for Disease Prevention and Control (ECDC) in 2015 [1], >100 cases of invasive cardiovascular infection and disseminated disease with Mycobacterium chimaera have been reported worldwide, not only in Europe, but also in North America and Asia [2, 3]. These infections are difficult to diagnose because of their nonspecific symptoms and specific microbiological requirements [4]. These infections were attributed to contamination from the heater-cooler units (HCUs) present in operating rooms as M. chimaera was found in their water tanks, probably transmitted on the surgical field through aerosolization when the exhaust fans were running [5]. Genomic studies on M. chimaera isolates showed, on the one hand, that most of the clinical isolates collected in different countries were clustered and, on the other hand, that many isolates cultured from the HCU water samples belong also to this cluster [6–8]. This raised the hypothesis of a common reservoir at the HCU factory. However, as these infections occurred over many years, the mean incubation being 21 months [2], and as M. chimaera is often present in water networks [9], all nosocomial infections might not have been caused by the factory contamination.
M. chimaera is a slow-growing nontuberculous mycobacterium (NTM) belonging to the Mycobacterium avium complex (MAC), now described as a subspecies of M. intracellulare [10]. It was rarely recognized as a pathogen before the outbreak and rarely identified at the species or subspecies level [11]. The purpose of the present study was to report the findings of the epidemiological, microbiological surveillance and molecular investigations conducted at health care facilities (HCFs) practicing open chest surgery in France. Clinical isolates were characterized for single nucleotide polymorphisms (SNPs) observed in comparison with HCU isolates. We also investigated the in vivo mutations that emerged between sequential isolates from the same patient.
METHODS
Epidemiological Investigations
Two investigations were organized targeting the 61 HCFs where cardiothoracic surgery was regularly performed under cardiopulmonary bypass (CPB) in France. The first, performed immediately after the ECDC June 2015 alert, sought clinical cases that could have occurred from January 1, 2010, to April 30, 2015 [12]. The second investigation, performed in 2017, sought additional cases from 2015 onwards and questioned HCU good practices including HCU microbiological analysis [13]. These investigations are detailed in the Supplementary Data and in Supplementary Tables 1 and 2.
M. chimaera Detection in Water Tank Samples of Heater-Cooler Units
A protocol adequate for water network samples was adapted from Radomski et al. [14] based on filtration and culture of specific media for mycobacteria research, as detailed in the Supplementary Data. Species and subspecies identification among the Mycobacterium avium complex was done using GenoType NTM-DR 1.0 (Bruker, Nehren, Germany) and ITS (internal transcribed spacers between 16S and 23S ribosomal DNA) or hsp65 gene PCR sequencing when necessary [11].
Whole-Genome Sequencing and SNP Analysis
DNA were extracted from M. chimaera solid cultures on Middlebrook 7H10 agar using the DNA Ultraclean Microbial kit (QIAGEN, Hilden, Germany). DNA libraries were prepared with the Nextera XT kit (Illumina, San Diego, CA, USA), and whole-genome sequencing (WGS) was performed with a MiSeq system (Illumina) and the MiSeq Reagent V2 (2×150) kit (Illumina). Sequence reads were aligned to the reference genome of M. chimaera strain ZUERICH-1 (sequence ID: NZ_CP015272.1 [7]) using Bionumerics, version 7.6 (Applied Maths, Gent, Belgium). The reads were trimmed excluding base call with a Phred score <15. The SNP signature was built using the strict-filtering (closed SNP set) option, retaining SNPs present in all the isolates, with a minimum coverage of 5X covered at least once in both forward and reverse directions, a minimum distance between the retained SNP position of 12 base pairs, and removing the nondiscriminatory position. The SNP matrix was used to build a maximum parsimony tree. Isolates were classified using the allele typing proposed by van Ingen et al. [7]. Genomic sequences are available from GenBank/NCBI in BioProject PRJNA667507.
RESULTS
M. chimaera Clinical Cases
Two cases (Patients P1 and P2) of disseminated M. chimaera disease were related to the outbreak out of 4 suspected cases reported by HCFs in the first investigation results. The 2 excluded cases were a patient with M. chimaera cultured from pericardial fluid but not submitted to CPB and a patient with a post-CPB infection but in whom the isolate was M. avium and not M. chimaera. P1 was a 61-year-old man who underwent cardiac surgery to replace part of his ascending aorta with a bio-prosthetic graft in 2012. He developed osteo-articular infection in 2014 (isolate P1a, Table 1), and a bloodstream infection was diagnosed 1 year later (isolate P1b) [2, 12]. He died 6 years later while being treated with a 4-antibiotic regimen, combining azithromycin, ethambutol, rifampicin, and moxifloxacin. P2 was a 53-year-old man who was diagnosed with disseminated granulomatous disease a few months after cardiac surgery involving a graft replacement of the aortic valve for aorta dilatation in 2009 and 2010 [12]. The patient presented M. chimaera–positive blood cultures 2 years after surgery and died shortly after. No invasive M. chimaera infection cases were reported after 2015.
Table 1.
Main Characteristics of the M. chimaera Clinical Isolates
| Characteristic | Patient 1 | Patient 2 | |
|---|---|---|---|
| Isolatea | P1a (I58) | P1b (I57) | P2 (I18) |
| Hospitala | H25 | H25 | H14 |
| SNP difference with strain Zuerich-1 | 5 | 10 | 5 |
| Clarithromycin MIC | 2 | 0.5 | 2 |
| Rifabutin MIC | 2 | 0.25 | 0.5 |
| Ethambutol MIC | 16 | 4 | 16 |
| Isoniazid MIC | >8 | 2 | 2 |
| Moxifloxacin MIC | 4 | 2 | 4 |
| Rifampicin MIC | >8 | 2 | 4 |
| Sulfamethoxazole/trimethoprim MIC | 8/152 | 1/19 | 8/152 |
| Amikacin MIC | 8 | 4 | 16 |
| Linezolid MIC | 32 | 4 | 16 |
| Ciprofloxacin MIC | 16 | 8 | 16 |
| Streptomycin MIC | 16 | 8 | 32 |
| Doxycycline MIC | 16 | >16 | >16 |
| Ethionamide MIC | 20 | 1.2 | 10 |
| Bedaquiline MIC | 0.25 | 0.25 | 0.25 |
MIC (in mg/L) was determined using the commercial microdilution method SLOMYCO Myco Sensititre (Thermo Scientific) following the manufacturer’s recommendations and CLSI guidance for bedaquiline testing [15].
Abbreviations: CLSI, Clinical and Laboratory Standards Institute; MIC, minimum inhibitory concentration; SNP, single nucleotide polymorphism.
aSee details in the Supplementary Table 4 list of isolates.
Contamination of HCU Water Samples With M. chimaera and HCU Practice Maintenance
Fifty-six HCFs (92% of all HCFs) answered the questionnaires, reporting the use of 227 HCUs. Specific measures to prevent infection were applied in 89% of these HCFs (50/56). Only 14% (8/56) systematically followed the manufacturer maintenance recommendations, and this dropped to 7% (4/56) for the new measures issued by Livanova in 2015 (eg, an increase in the frequency of water replacement and disinfection of the water circuit) (Supplementary Table 2).
HCFs declared that 41 (23%) of the 175 HCUs cultivated for MAC were positive. Our national reference laboratory received 75 additional HCU water samples from 11 HCFs, of which 36 (48%) were positive for various species of mycobacteria (Supplementary Table 3, Supplementary Figure 1). Out of these 36 positive HCU samples, 24 were positive for M. chimaera at a mean quantification (range) of 104 CFU/L (101–105/L). HCUs were also positive for some other mycobacteria: 17 for M. chelonae (mean, 3.3 × 103 CFU/L), 8 for M. mucogenicum (mean, 1.1 × 103 CFU/L), 8 for M. gordonae/paragordonae (mean, 3.4 × 103 CFU/L), 2 for M. avium (mean, 8 × 103 CFU/L), and 2 for M. peregrinum (mean, 1.2 × 103 CFU/L).
Genomic Comparison of M. chimaera Isolates
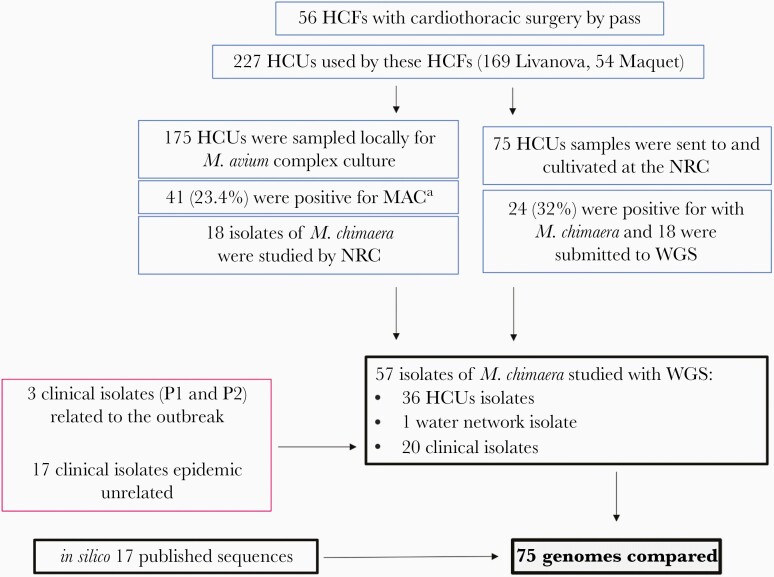
A total of 57 M. chimaera isolates (detailed in Figure 1 and Supplementary Table 4) were submitted to WGS analysis: the 3 clinical isolates obtained from the 2 patients (P1a, P1b, P2), 36 isolates from HCU water samples of 11 HCFs, 17 clinical M. chimaera isolates a priori unrelated to the outbreak (from 2 myocardial biopsies and 1 breast biopsy and 14 sputum), and 1 environmental isolate from the water supply network of a French hospital. WGS data were compared with the genome sequence of the epidemic strain M. chimaera ZUERICH-1 [7]. Sixteen other genomes published for clinical cases described in the United States (n = 13) [16] and in Ireland (n = 3) were added to the comparison [17].
Figure 1.
Overview of studied Mycobacterium chimaera isolates and their origin according to epidemiological investigations. Abbreviations: HCFs, health care facilities; HCU, heater-cooler unit; MAC, Mycobacterium avium complex; WGS, whole-genome sequencing. *Specific identification was not performed systematically for M. chimaera.
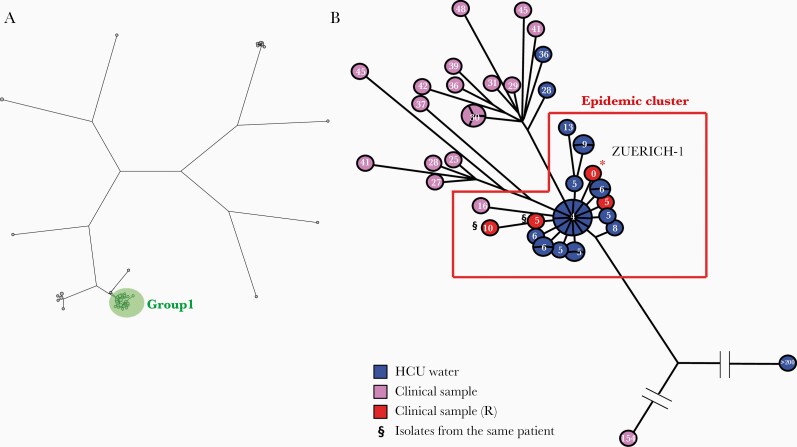
Genomic comparison analysis generated 55 638 SNPs, which were used to build a maximum parsimony tree, shown in Figure 2A. The genomes were distributed among 9 groups, with 52 isolates clustering with the epidemic strain M. chimaera ZUERICH-1, namely group 1, in which <100 SNP differences were observed between the sequences [7]. A subsequent analysis of these 52 genomes generated 694 SNPs, which were used to build a second maximum parsimony tree, shown in Figure 2B. This showed more precisely 30 isolates clustering with the epidemic strain M. chimaera ZUERICH-1, with a mean pairwise distance of 4 SNPs. The 3 isolates corresponding to the French clinical cases (P1 and P2 isolates) were confirmed to belong to the epidemic cluster, as well as 26 out of 36 (72%) M. chimaera isolates isolated from HCUs used in French hospitals. All were Livanova HCUs, and none were from Maquet ones. Clinical isolates were typed according to an allele classification proposed by van Ingen et al. and belonged to subgroup 1.1 (Supplementary Table 4) [7]. One isolate (#8) obtained from a sputum sample of a nonrelated patient with no history of cardiac surgery was unexpectedly phylogenetically linked to the epidemic cluster but with 16 SNP differences and belonged to subgroup 1.8.
Figure 2.
Phylogenetic analysis of Mycobacterium chimaera isolates. A, Maximum parsimony tree (logarithmically scaled) built based on the 55 638 SNP positions found through comparison of the 75 genomes that were mapped to the reference genome (*) of M. chimaera ZUERICH-1. The 52 isolates, including Zuerich-1, belonging to group 1 are gathered in a green circle. B, Maximum parsimony tree (logarithmically scaled) built based on the 694 SNP positions found through comparison of the genomes that were mapped to the reference genome of M. chimaera ZUERICH-1. The number of SNP differences with regard to the epidemic strain M. chimaera ZUERICH-1 is indicated inside each circle. Isolates from HCU water samples are labeled in blue, isolates from clinical samples in purple, and isolates from clinical samples related to the outbreak in red. §Two isolates isolated from the same patient. Abbreviations: HCU, heater-cooler unit; SNP, single nucleotide polymorphism.
Specific Analyses of Clinical Isolates
We specifically analyzed the genomes of the 3 clinical isolates from the 2 cases, detailing the SNPs observed with regards to the epidemic strain Zuerich-1, as presented in Table 2. Among 11 SNPs, all were located in a protein-coding sequence, and only 1 was synonymous. Minimum inhibitory concentrations (MICs) of several antimicrobial agents were determined for the clinical isolates; these are shown in Table 1.
Table 2.
SNP Analyses Determined From the Genomic Comparison of Clinical Isolates From Patients P1 and P2 and of Environmental Isolates Related to the P2 Case
| Isolates | Positiona | Nucleotide Modification | Gene Characterization | Codon | Mutation Effect |
|---|---|---|---|---|---|
| Comparison of clinical isolates from the P1 and P2 cases | |||||
| P1a, P1b, P2 | 2 587 843 | T->C | Glycosyltransferase | CAC->CGC | H->R |
| P1a, P1b, P2 | 3 709 626 | G->A | Type III polyketide synthase | GCG->GTG | A->V |
| P1a, P1b, P2 | 4 520 250 | T->A | Signal peptidase I | GAA->GAT | E->D |
| P1a, P1b, P2 | 4 919 479 | A->G | Helix-turn-helix transcriptional regulator | TCC->CCC | S->P |
| P1a, P1b | 2 67 758 | C->A | Hypothetical protein | GAC->TAC | D->Y |
| P1b | 2 22 733 | T->C | Decaprenyl-phosphate phosphoribosyltransferase | TTC->CTC | Synonymous |
| P1b | 1 256 515 | G->T | IS481 family transposase | AGC->ATC | S->I |
| P1b | 1 256 515b | G->T | NAD(P)-dependent oxidoreductase | AGC->AGA | S->R |
| P1b | 1 509 107 | C->G | LuxR family transcriptional regulator | CGC->GGC | R->G |
| P1b | 4 708 684 | G->A | RND family transporter (mmpL5) | TGG->TGA | W->UGA |
| P1b | 5 517 224 | T->A | Porphobilinogen synthase | ATG->TTG | M->L |
| P2 | 2 436 71 | T->C | Arabinosyltransferase | ATG->GTG | M->V |
| Comparison of environmental isolates related to the P2 case | |||||
| I36, I35 | 5 234 848 | A->G | FAD-dependent oxidoreductase | CAC->CGC | H->R |
| I36, I35 | 5 947 828 | C->A | NAD(P)/FAD-dependent oxidoreductase | GAC->AAC | D->K |
| I35 | 3 350 727 | A->T | PPE family protein | CTG->CAG | L->Q |
| I30, I34 | 1 367 077 | T->C | Hypothetical protein | GTG->GCG | V->A |
| I30, I34 | 2 057 124 | C->A | Valine-tRNA ligase | CCC->CCA | Synonymous |
| I30, I34 | 5 952 067 | G->C | Cytochrome P450 | CAC->CAG | H->Q |
| I30, I34 | 5 952 114 | G->T | NIPSNAP family protein | CTC->CTA | Synonymous |
| I30, I34 | 6 159 327 | G->A | Hypothetical protein | CGC->CAC | R->H |
| I30 | 539 741 | C->A | SpoIIE family protein phosphatase | CGT->CTG | R->L |
| I30 | 576 340 | C->T | Type II secretion system F family protein | CTG->TAG | L->stop |
| I30 | 4 243 378 | C->G | Hypothetical protein | GGG->GGC | Synonymous |
| I34 | 1 204 266 | G->T | LuxR family transcriptional regulator | GCC->GCA | Synonymous |
| I34 | 1 562 307 | G->A | Alpha/beta hydrolase | CAG->CAA | Synonymous |
| I34 | 2 093 313 | C->T | PE-PPE domain–containing protein | GCG->GCA | Synonymous |
| I34 | 2 447 021 | T->C | DNA primase | ATC>ACC | I->T |
| I34 | 2 842 316 | C->A | MCE family protein | CTG>CTT | Synonymous |
| I34 | 4 179 540 | T->C | Hypothetical protein | ACA>ACG | Synonymous |
| I34 | 4 243 378 | C->G | Hypothetical protein | GGG>GGC | Synonymous |
| I34 | 5 019 553 | G->A | Unknown function | CCC>CCT | Synonymous |
aSNPs were generated with regards to the sequence of the epidemic strain Mycobacterium chimaera ZUERICH-1 (NCBI reference sequence NZ_CP015272.1).
bThe SNP at the 1256515 position affected 2 genes, 1 forward and the other in reverse.
Between the 2 sequential isolates from patient P1, P1a isolates in 2014 and P1b isolates in 2015, 5 SNPs were observed. One SNP produced a STOP codon in the gene MYCOZU1_RS21880. This gene contains a conserved domain belonging to the mycobacterial membrane protein large (mmpL) family, a resistance-nodulation-cell division (RND) family transporter reported to be involved in multisubstrate transport; it is known to contribute to virulence and pathogenicity [18]. MYCOZU1_RS21880 exhibits a 67.8% similarity at the nucleotide level with M. tuberculosis mmpL5 (Supplementary Table 5). However, the M. chimaera mmpL5 ortholog would more likely correspond to the gene MYCOZU1_RS13900, which presents a higher similarity (76.2%) and is found in synteny with an mmpS-like gene (MYCOZU1_RS13905) and a tet-R-like gene (MYCOZU1_RS13910) upstream [19]. This SNP was observed in the genome of the P1b isolate, which was isolated after 1 year of antibiotic treatment including azithromycin, ethambutol, rifampicin, and moxifloxacin.
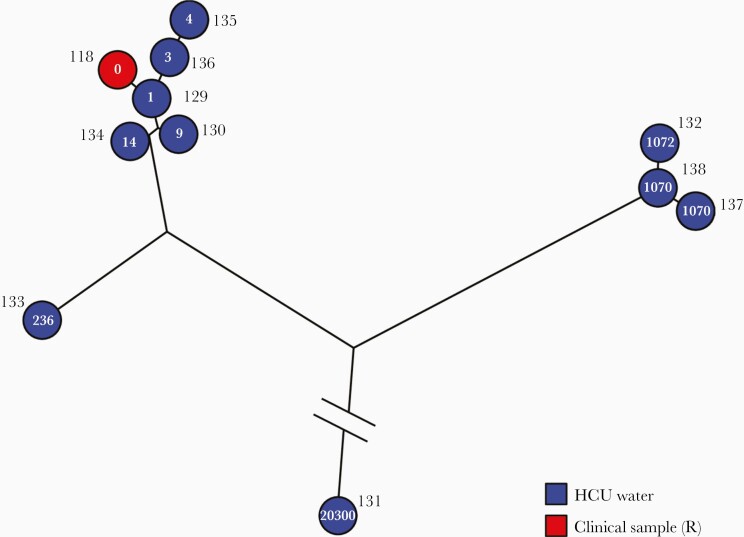
No HCU isolates were obtained from the hospital where P1 was operated on, as the samples were declared negative for many months after the case was declared. P2 was operated on in hospital H5 and diagnosed in hospital H14. Fourteen water samples were recovered from this hospital, and 9/14 (64%) were positive for M. chimaera. In total, 10 HCU isolates were sequenced. Genomic comparison of P2-related isolates (clinical and environmental) is shown in Figure 3. Five HCU isolates showed >100 SNP differences and thus were not related to the outbreak. Five HCU isolates belonged to the epidemic cluster, showing 1–14 SNP differences from the P2 isolate sequence; these are presented in Table 2. Half of these SNPs were synonymous.
Figure 3.
Phylogenetic analysis of Mycobacterium chimaera isolates related to P2. Maximum parsimony tree (logarithmically scaled) built based on the 21 106 SNP positions mapped to the genome of M. chimaera ZUERICH-1 found through comparison of the 11 isolates related to patient P2. The number of SNP differences compared with patient P2’s isolate is indicated inside each circle. Isolates from HCU water samples are labeled in blue, and isolates from clinical samples related to the outbreak are labeled in red. Abbreviations: HCU, heater-cooler unit; SNP, single nucleotide polymorphism.
DISCUSSION
Contamination of HCU water cooling systems by M. chimaera has led to a worldwide outbreak among patients undergoing cardiopulmonary bypass surgery [2]. In this study, we report the results of investigations and surveillance conducted in France, focusing on the results of genomic analyses of M. chimaera isolates obtained from patients and HCU contamination related to the outbreak. Although most of the isolates clustered with the outbreak strain Zuerich-1, some did not, and within the cluster several SNP differences resulting in substitutions in gene-coding regions were observed.
In the search for clinical cases related to the outbreak in all the medical facilities performing open chest surgery with cardiopulmonary bypass in France, only 2 cases were declared, which is far fewer than in other European countries (eg, 18 cases in the United Kingdom [6], 6 cases in Switzerland [20]). This could be due to a shorter period of investigation (5 years vs 8 and 9 years, respectively) or differences in practices. Another explanation could be under-reporting or underdiagnosis of cases as not all HCFs are searching for M. chimaera infections in re-operated patients in infectious contexts (10% of cases in France). However, cases were also sought through an active network of cardiologists, surgeons, and microbiologists which regularly surveyed infective endocarditis without further new cases detected [21]. The majority of HCFs declared, however, having implemented prevention measures and maintenance protocols to reduce exposure of patients to HCUs, so even if compliance to manufacturer recommendations was not complete or was loosely adapted to the reality of practice, this could explain the small number of cases despite the positivity of many HCU water samples.
The manufacturers revised their recommendations after the ECDC alert, and the recommendations were difficult to apply. For example, the daily exchange of total water volume from the reservoir was not possible due to the architecture of the water tank, and the exchange requires too much personal time. The HCF personnel and staff, however, performed disinfection with the elements and materials they had and as frequently as possible. It was eventually shown that the HCUs were mainly contaminated during their production at the factory, and consequently the disinfection measures recommended were mainly useful to decrease the M. chimaera load but not prevent contamination of the water contained in the HCUs [4].
Following a protocol standardized for mycobacterial detection and applied to HCU water samples, we showed that M. chimaera was the most frequently isolated and abundant mycobacteria. MAC are waterborne, and M. chimaera can be recovered from various environments: drinking water distribution systems, household water, and associated biofilms [9]. MAC can be found at levels up to 106 UFC/L in water distribution systems [22]. Strikingly, slow-growing mycobacteria were more prevalent in HCU water than rapid-growing mycobacteria, contrary to what is observed in the water supply network in France [23]. Other mycobacterial species were also detected in HCU water [24], and their presence in the operating room during cardiac surgery is a potential nosocomial threat. Infection with M. abscessus [25] and M. wolinskyi [26] linked to contaminated HCUs was indeed previously reported, and M. chelonae was one of the mycobacterial species most frequently found in endocarditis [27]. M. chimaera HCU contamination could also put health care workers at risk of developing pneumonia [28].
WGS and SNP analysis have become the reference method for the molecular epidemiological study of M. tuberculosis [29], but few studies on NTM are available. WGS has been used to connect M. avium isolated in household water to respiratory disease, showing a maximum distance of 51 SNPs between respiratory and household isolates [9]. Concerning the M. chimaera outbreak, several WGS studies compared clinical and HCU isolates and showed a clonal signature with a mean pairwise distance ranging from 4 to 10 SNP differences [6–8]. Using the same methodology, we found very low diversity in the epidemic cluster for the M. chimaera isolates in France, with a mean pairwise distance of 4 SNPs, and confirmed that the 2 suspected patients were infected by the M. chimaera epidemic strain. In our study, 72% of M. chimaera found in HCU water samples belonged to the epidemic group cluster. Other studies have also found a predominance of the epidemic clone, with levels ranging from 48% [6] to 96% of the studied isolates [8]. Surprisingly, 1 isolate not related to the outbreak was shown to belong to the epidemic cluster. This was also observed in another study in the United Kingdom that identified 2 isolates from control patients as belonging to the epidemic cluster [6]. Bioinformatic analysis is a major source of analysis variation that can affect SNP variant calling [29]. WGS analysis is a powerful, discriminative tool, but we do not know its exact specificity.
Our study has several limitations. Case research and HCU surveillance were not systematic but based on declaration and retrospective research, as the mycobacteriology laboratories in France are not centralized like in the United Kingdom, for instance [30]. All the HCU isolates were sampled after 2015, although contamination occurred a few years before. There were indeed several years’ delay between patient contamination and microbial analysis of HCU water samples [6–8]. Lastly, as we decontaminated the water sample, we cannot know about contamination by bacteria other than mycobacteria, as was reported by Chand et al. [6].
Specific SNP analysis of clinical and HCU isolates revealed diversity even for those included in the epidemic cluster. Interestingly, SNPs observed in the clinical isolates showed very few nonsynonymous mutations, compared with SNPs observed in the HCU isolates. This could suggest that only SNPs conferring an adaptive advantage (survival, virulence) are selected during in-host evolution. Experimental studies have shown that only a few mutations can increase virulence. In a follow-up study on M. avium respiratory diseases, the genome sequences of intrapatient isolates were highly similar, with only 0–19 SNP differences, but exhibited increased virulence features compared with the first isolates [31]. Patient P1b’s isolate showed a nonsense mutation in an MmpL protein that was among the additional mmpL genes found in M. chimaera but not M. tuberculosis [32] and exhibited homology with mmpL5. MmpL5 protein is an RND superfamily efflux pump involved in siderophores and the export of drugs such as azole and bedaquiline [19]. The P1b mmpL5 mutated isolate showed a decreased MIC value compared with the parent strain P1 isolated 1 year before. In M. tuberculosis, mmpL5 was shown to be a nonessential gene in the presence of heme and hemoglobin as an iron source [33]. Further studies are required to determine if this mutation is linked to an increase in virulence or fitness.
CONCLUSIONS
In conclusion, we have presented a complete epidemiological investigation of M. chimaera infection associated with heater-cooler units in France through practice assessment, microbiological surveillance, case detection, and molecular analysis. Two cases were reported and confirmed by whole-genome analysis. Due to the high prevalence of the M. chimaera epidemic strain found in HCUs, regulatory action and continuous surveillance are still necessary.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We thank the persons who participated in collecting isolates and sending them to the CNR-MyRMA: Corentine Alauzet, Laurent Cavalié, Christine Chefson-Girault, Olivier Belmonte, Catherine Bounhiol Colin, Anne Bourgoin, Anne Bousseau, Lucien Brasme, Céline Cépoulain, Florence Cizeau, Christophe De Champs, Jean-Winoc Decousser, David Ducellier, Marguerite Fines Guyon, Sandra Fournier, Karine Gambarotto, Marion Grare, Hélène Guet-Revillet, Chloe Jansen, Frederic Janvier, Najiby Kassis-Chikhani, Valerie Lalande, Philippe Lanotte, Annick Lefebvre, Claire Lesteven, Marc Levy, Thomas Maitre, Sandra Malavaud, Stéphanie Morel, Isabelle Podglajen, Elodie Poisnel, Cécile Poulain, Marie-Fleur Rafidiarisoa, Jean Ringeval, Alexandre Rivier, Jerome Robert, Christine Roques-Ceschin, Soumaya Skalli, Ousmane Traore, Celine Vauterin, and Charlotte Verdet. We thank Diana Machado and Wladimir Sougakoff for our discussion about the efflux pump. We thank Thomas Kohl for his analysis and discussion on M. chimaera genomes.
We thank also the technicians of Lariboisière Bacteriology Laboratory for their expert technical assistance: Christine Bisilliat-Gardet, Véronique Charlier, Marie-Emmanuelle Hemet, Marilyne Lemaire, Isabelle Lacrampe, Patricia Lawson-Body, Marie Monjean, Fabienne Meunier, Sylvie Tenza, Odile Vissouarn, and all other members of the laboratory.
Collaborators. Alexandra Aubry, Isabelle Bonnet, Gauthier Pean de Ponfilly, Hervé Jacquier, Vincent Jarlier, Florence Morel, Jerome Robert, Wladimir Sougakoff, Nicolas Veziris.
Financial support. This work was supported by an annual grant from Santé Publique France (SPF) and Direction Générale de l’Offre de Soins (DGOS) for the associate laboratory of the National Reference Center for Mycobacteria and Antimycobacterial Resistance (CNR-MyRMA).
Potential conflicts of interest. The authors report no conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Author contributions. E.L., C.D., F.M., S.F., G.L., J.R., A.C., and E.C. collected epidemiological data. E.L., S.K., F.M., K.L., and H.B. provided microbiological and genomic data. E.L. and E.C. wrote the first draft of the manuscript, which was revised by all other authors.
Patient consent. Written informed consent was obtained from the patient’s next of kin. We followed procedures in accordance with the Helsinki Declaration. Information was anonymized. According to the French regulation on observational database analyses including microbial database, the study did not have specific ethical requirements.
Contributor Information
CNR-MyRMA:
Corentine Alauzet, Laurent Cavalié, Christine Chefson-Girault, Olivier Belmonte, Catherine Bounhiol Colin, Anne Bourgoin, Anne Bousseau, Lucien Brasme, Céline Cépoulain, Florence Cizeau, Christophe De Champs, Jean-Winoc Decousser, David Ducellier, Marguerite Fines Guyon, Sandra Fournier, Karine Gambarotto, Marion Grare, Hélène Guet-Revillet, Chloe Jansen, Frederic Janvier, Najiby Kassis-Chikhani, Valerie Lalande, Philippe Lanotte, Annick Lefebvre, Claire Lesteven, Marc Levy, Thomas Maitre, Sandra Malavaud, Stéphanie Morel, Isabelle Podglajen, Elodie Poisnel, Cécile Poulain, Marie-Fleur Rafidiarisoa, Jean Ringeval, Alexandre Rivier, Jerome Robert, Christine Roques-Ceschin, Soumaya Skalli, Ousmane Traore, Celine Vauterin, and Charlotte Verdet
References
- 1. European Centre for Disease Prevention and Control. Invasive Cardiovascular Infection by Mycobacterium chimaera – 30 April 2015. Stockholm: European Centre for Disease Prevention and Control;2015. [Google Scholar]
- 2. Lecorche E, Pean de Ponfilly G, Mougari F, et al. . Disseminated Mycobacterium chimaera following open-heart surgery, the heater-cooler unit worldwide outbreak: case report and minireview. Front Med (Lausanne) 2020; 7:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Riccardi N, Monticelli J, Antonello RM, et al. . Mycobacterium chimaera infections: an update. J Infect Chemother 2020; 26:199–205. [DOI] [PubMed] [Google Scholar]
- 4. Hasse B, Hannan MM, Keller PM, et al. ; M. chimaera ISCVID Investigators and; ISCVID Executive Committee; Infectious Diseases Specialists; Hospital Epidemiologists; Microbiologists and Molecular Typing Specialists; Cardiac Surgeons/Perfusionists/Cardiologists; Ophthalmology; Anaesthesiologists; Public Health. International Society of Cardiovascular Infectious Diseases guidelines for the diagnosis, treatment and prevention of disseminated Mycobacterium chimaera infection following cardiac surgery with cardiopulmonary bypass. J Hosp Infect 2020; 104:214–35. [DOI] [PubMed] [Google Scholar]
- 5. Schreiber PW, Sax H. Mycobacterium chimaera infections associated with heater-cooler units in cardiac surgery. Curr Opin Infect Dis 2017; 30:388–94. [DOI] [PubMed] [Google Scholar]
- 6. Chand M, Lamagni T, Kranzer K, et al. . Insidious risk of severe Mycobacterium chimaera infection in cardiac surgery patients. Clin Infect Dis 2017; 64:335–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van Ingen J, Kohl TA, Kranzer K, et al. . Global outbreak of severe Mycobacterium chimaera disease after cardiac surgery: a molecular epidemiological study. Lancet Infect Dis 2017; 17:1033–41. [DOI] [PubMed] [Google Scholar]
- 8. Hasan NA, Epperson LE, Lawsin A, et al. . Genomic analysis of cardiac surgery-associated Mycobacterium chimaera infections, United States. Emerg Infect Dis 2019; 25:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lande L, Alexander DC, Wallace RJ Jr, et al. . Mycobacterium avium in community and household water, suburban Philadelphia, Pennsylvania, USA, 2010-2012. Emerg Infect Dis 2019; 25:473–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Ingen J, Turenne CY, Tortoli E, et al. . A definition of the Mycobacterium avium complex for taxonomical and clinical purposes, a review. Int J Syst Evol Microbiol 2018; 68:3666–77. [DOI] [PubMed] [Google Scholar]
- 11. Lecorche E, Haenn S, Mougari F, et al. ; CNR-MyRMA. Comparison of methods available for identification of Mycobacterium chimaera. Clin Microbiol Infect 2018; 24:409–13. [DOI] [PubMed] [Google Scholar]
- 12. Lebreton G, Fournier S, Subiros M, et al. . Risques infectieux liés aux générateurs thermiques utilisés pendant les circulations extracorporelles: info ou intox? JCTCV 2017; 21. [Google Scholar]
- 13. Daniau C, Lecorche E, Mougari F, et al. . Infections invasives à Mycobacterium chimaera en chirurgie cardiaque et évaluation des pratiques autour des matériels de circulation extracorporelle. Hygiènes 2019; 27:351–9. [Google Scholar]
- 14. Radomski N, Cambau E, Moulin L, et al. . Comparison of culture methods for isolation of nontuberculous mycobacteria from surface waters. Appl Environ Microbiol 2010; 76:3514–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Woods GL, Brown-Elliott BA, Conville PS. Susceptibility Testing of Mycobacteria, Nocardiae, and Other Aerobic Actinomycetes [Internet]. 2nd ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2011 March, Report No. M24-A2. [PubMed] [Google Scholar]
- 16. Epperson LE, Timke M, Hasan NA, et al. . Evaluation of a novel MALDI Biotyper algorithm to distinguish Mycobacterium intracellulare from Mycobacterium chimaera. Front Microbiol 2018; 9:3140.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mac Aogáin M, Roycroft E, Raftery P, et al. . Draft genome sequences of three Mycobacterium chimaera respiratory isolates. Genome Announc 2015; 3:e01409–15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Machado D, Lecorche E, Mougari F, et al. . Insights on Mycobacterium leprae efflux pumps and their implications in drug resistance and virulence. Front Microbiol 2018; 9:3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Briffotaux J, Huang W, Wang X, Gicquel B. MmpS5/MmpL5 as an efflux pump in Mycobacterium species. Tuberculosis (Edinb) 2017; 107:13–9. [DOI] [PubMed] [Google Scholar]
- 20. Sax H, Bloemberg G, Hasse B, et al. . Prolonged outbreak of Mycobacterium chimaera infection after open-chest heart surgery. Clin Infect Dis 2015; 61:67–75. [DOI] [PubMed] [Google Scholar]
- 21. Limonta S, Cambau E, Erpelding ML, et al. ; EI 2008 de l’AEPEI working group. Infective endocarditis related to unusual microorganisms: a prospective population-based study. Open Forum Infect Dis 2020; 7:ofaa127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Falkinham JO 3rd, Norton CD, LeChevallier MW. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other mycobacteria in drinking water distribution systems. Appl Environ Microbiol 2001; 67: 1225–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Delafont V, Mougari F, Cambau E, et al. . First evidence of amoebae-mycobacteria association in drinking water network. Environ Sci Technol 2014; 48:11872–82. [DOI] [PubMed] [Google Scholar]
- 24. Kaelin MB, Kuster SP, Hasse B, et al. . Diversity of nontuberculous mycobacteria in heater-cooler devices - results from prospective surveillance. J Hosp Infect 2020; S0195-6701(20)30105-5. [DOI] [PubMed] [Google Scholar]
- 25. Baker AW, Lewis SS, Alexander BD, et al. . Two-phase hospital-associated outbreak of Mycobacterium abscessus: investigation and mitigation. Clin Infect Dis 2017; 64:902–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nagpal A, Wentink JE, Berbari EF, et al. . A cluster of Mycobacterium wolinskyi surgical site infections at an academic medical center. Infect Control Hosp Epidemiol 2014; 35:1169–75. [DOI] [PubMed] [Google Scholar]
- 27. Shi-Min Y. Mycobacterial endocarditis: a comprehensive review. Rev Bras Cir Cardiovasc 2015; 30:93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rosero CI, Shams WE. Mycobacterium chimaera infection masquerading as a lung mass in a healthcare worker. IDCases 2019; 15:e00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meehan CJ, Goig GA, Kohl TA, et al. . Whole genome sequencing of Mycobacterium tuberculosis: current standards and open issues. Nat Rev Microbiol 2019; 17:533–45. [DOI] [PubMed] [Google Scholar]
- 30. Scriven JE, Scobie A, Verlander NQ, et al. . Mycobacterium chimaera infection following cardiac surgery in the United Kingdom: clinical features and outcome of the first 30 cases. Clin Microbiol Infect 2018; 24:1164–70. [DOI] [PubMed] [Google Scholar]
- 31. Kannan N, Lai Y-P, Haug M, et al. . Genetic variation/evolution and differential host responses resulting from in-patient adaptation of Mycobacterium avium. Infect Immun 2019; 87:e00323–18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Viljoen A, Dubois V, Girard-Misguich F, et al. . The diverse family of MmpL transporters in mycobacteria: from regulation to antimicrobial developments. Mol Microbiol 2017; 104:889–904. [DOI] [PubMed] [Google Scholar]
- 33. Zhang L, Hendrickson RC, Meikle V, et al. . Comprehensive analysis of iron utilization by Mycobacterium tuberculosis. PLoS Pathog 2020; 16:e1008337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.