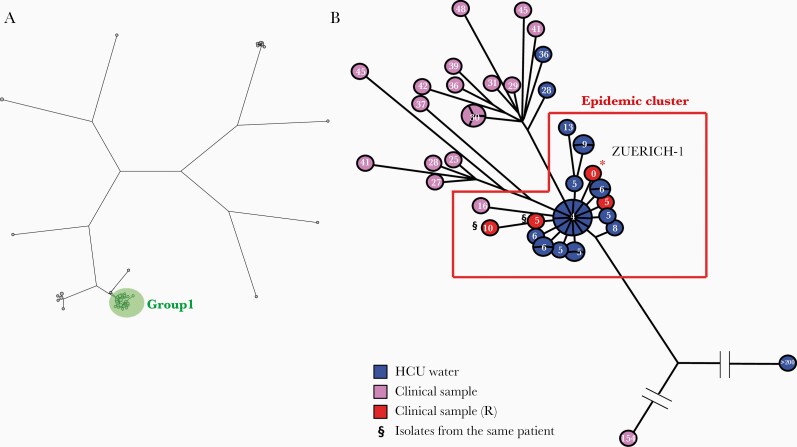
Figure 2.
Phylogenetic analysis of Mycobacterium chimaera isolates. A, Maximum parsimony tree (logarithmically scaled) built based on the 55 638 SNP positions found through comparison of the 75 genomes that were mapped to the reference genome (*) of M. chimaera ZUERICH-1. The 52 isolates, including Zuerich-1, belonging to group 1 are gathered in a green circle. B, Maximum parsimony tree (logarithmically scaled) built based on the 694 SNP positions found through comparison of the genomes that were mapped to the reference genome of M. chimaera ZUERICH-1. The number of SNP differences with regard to the epidemic strain M. chimaera ZUERICH-1 is indicated inside each circle. Isolates from HCU water samples are labeled in blue, isolates from clinical samples in purple, and isolates from clinical samples related to the outbreak in red. §Two isolates isolated from the same patient. Abbreviations: HCU, heater-cooler unit; SNP, single nucleotide polymorphism.