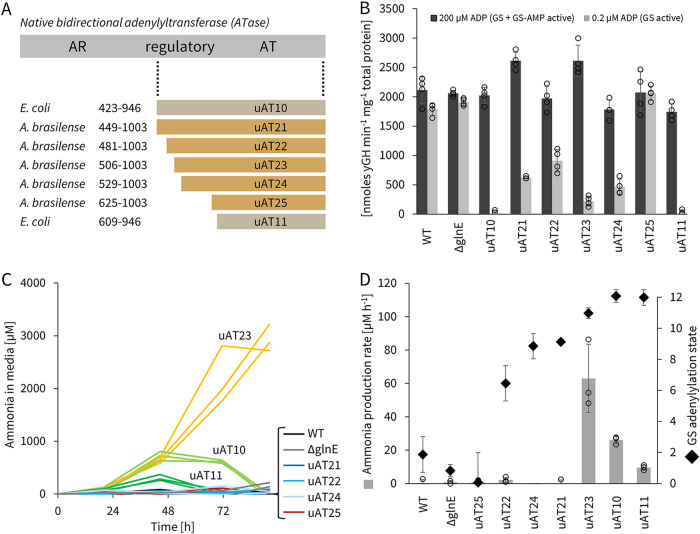
FIG 2.
GS activity can be modulated by uATs, and GS shutdown leads to ammonia release. (A) Alignment of 7 truncations of ATase from either E. coli and A. brasilense native ATase genes (glnE) that were tested for uAT activity. AT and AR denote the adenylyl-transferring and the adenylyl-removing domains, respectively. Average glutamine synthetase (GS) activity (B) and ammonia concentration in medium (C) of constitutively uAT-expressing A. brasilense ΔglnE strains inoculated at an OD600 of 0.1 in semisolid NFbHP medium (circuit R2 from pTS7 [Fig. S21]). (B) GS activity was measured by γ-glutamyl hydroxamate (γGH) production rates at high (200 μM) and low (0.2 μM) ADP concentrations (see Fig. S3A and B and S4 for the assay); rates are normalized to total protein content determined by the Bradford assay. (C) Ammonia was quantified by the indophenol method (see Fig. S6 for the assay); error bars are for triplicate biological replicates. (D) Comparison of maximal ammonia production rates observed between 20 and 70 h from data in panel C to computed GS adenylylation states (see Fig. S3C for calculations); error bars are standard deviations of biological replicates shown in panels B and C.