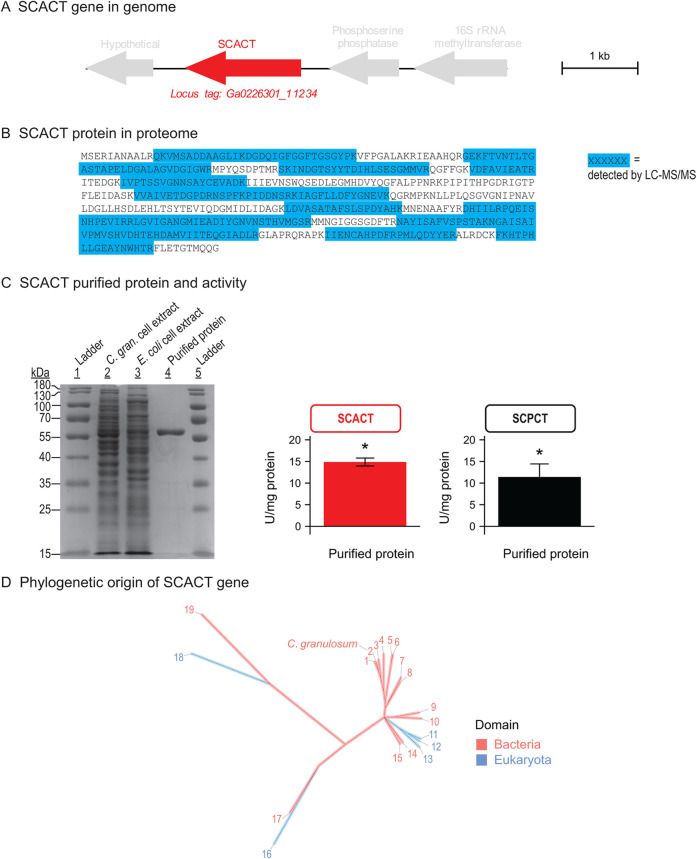
FIG 4.
The gene encoding SCACT in C. granulosum is a bacterial homolog, pointing to a bacterial origin of the SCACT/SCS pathway. (A) Location of the SCACT gene in the genome of C. granulosum. (B) Detection of SCACT protein in the proteome of C. granulosum. Specific peptides detected by LC-MS/MS are highlighted in the full protein sequence. Peptides shown were detected in each of two biological replicates (batches of protein prepared from independent cultures). (C) Production and enzymatic activity of recombinant SCACT protein in E. coli. Purity shown by SDS-PAGE was 92%. The amounts of protein loaded were 7.7, 5.8, and 0.8 μg for lanes 2, 3, and 4. Enzymatic activity is mean ± standard error of 3 biological replicates. Means different from 0 (P < 0.05) are marked with an asterisk. (D) Phylogenetic tree of SCACT protein sequences, showing that SCACT in C. granulosum is a bacterial homolog. Proteins included are those with experimental evidence of enzymatic activity. Protein sequences are as follows: 1, Acidipropionibacterium acidipropionici; 2, Cutibacterium granulosum; 3, Propionibacterium freudenreichii; 4, Corynebacterium diphtheriae; 5, Acetobacter cerevisiae; 6, Acetobacter aceti; 7, Snodgrassella alvi; 8, Moraxella catarrhalis; 9, Bacteroides fragilis; 10, Escherichia coli (homolog 1); 11, Aspergillus nidulans; 12, Saccharomyces cerevisiae; 13, Trichomonas vaginalis; 14, Acinetobacter baumannii; 15, Clostridium kluyveri; 16, Fasciola hepatica; 17, Yersinia pestis; 18, Trypanosoma brucei (homolog 1); and 19, Ralstonia eutropha. Full information on proteins is in Data Set S2. Abbreviations: C. gran., Cutibacterium granulosum; SCACT, succinyl-CoA:acetate CoA-transferase; SCPCT, succinyl-CoA:propionate CoA-transferase; SCS, succinyl-CoA synthetase (ADP forming).