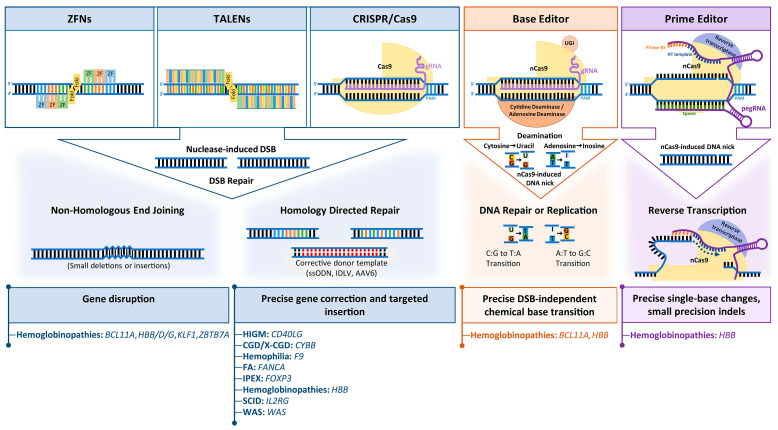
Figure 2.
Structure, function and HSC-based application of common gene-editing platforms. ZFNs, TALENs and CRISPR/Cas9 are chimeric proteins comprising customizable sequence-specific DNA binding domain (e.g., zinc finger—ZF, transcription activator-like effector proteins—TALEs or single guide RNA—sgRNA) and a nonspecific nuclease that mediates DNA cleavage (e.g., FokI nuclease in the context of ZFNs and TALENs and Cas9 in the case of CRISPR/Cas9). DNA double-strand breaks generated by ZFNs, TALENs and CRISPR/Cas9 are mainly repaired via two endogenous pathways: (1) error-prone non-homologous end joining (NHEJ), which occurs throughout the cell cycle and corrects breaks through ligation of DNA ends, or (2) by precise homology-directed repair (HDR), in the presence of donor template provided, e.g., as synthetic single-stranded oligodeoxynucleotides (ssODNs), insertion-defective lentiviral vector (IDLV) or adeno-associated virus 6 vector (AAV6) components,. Base editors are chimeric proteins composed of a mutated nuclease, such as Cas9 nickase (nCas9), a catalytic domain capable of deaminating a cytidine or adenine base to induce transition mutations, and a uracil glycosylase inhibitor to prevent base excision repair of the transition event. Prime editors are chimeric proteins exploiting an extended gRNA, termed prime editing guide RNA (pegRNA), and a nCas9 fused to a reverse transcriptase, which nick the DNA to allow pegRNA binding of flanking gRNA to serve as primer of pegRNA-directed reverse transcription of the desired sequence change.