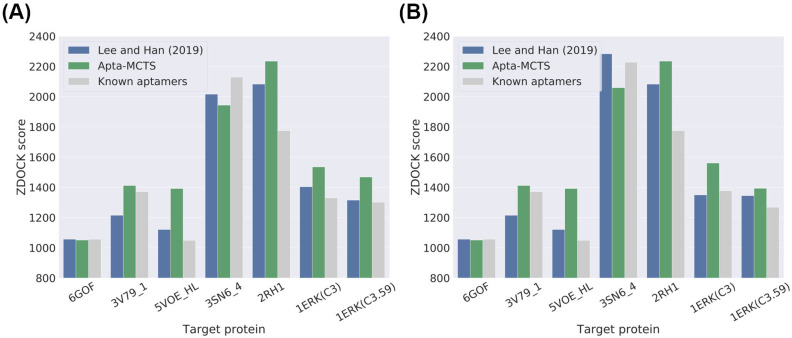
Fig 3. Evaluation of aptamer sequence generation in terms of binding affinity with six target proteins using ZDOCK docking simulation.
(A) ZDOCK scores of aptamers using protein structures from the PDB and (B) ZDOCK scores using structures built via the Swiss-Model server. For protein 1ERK, there are two known aptamers, C3 and C3.59, with 90 and 59 bases, respectively. We compared the docking scores of aptamer sequences generated by our model (in green bar) and Lee et al. [21] (in blue); the known aptamers (in gray) are listed in Table 2. The candidates of our Apta-MCTS (in green bar) yielded higher docking scores than the results reported by Lee et al. [21] (in blue) and the known aptamers (in gray) for five cases: 3V79_1, 5VOE (chain H and L), 2RH1, 1ERK(C3) and 1ERK(C3.59). Additional details are available in S2 Table for (A) and S3 Table for (B).