Fig. 6. Lysine methylation blocks the recruitment of autophagy receptors to R. parkeri.
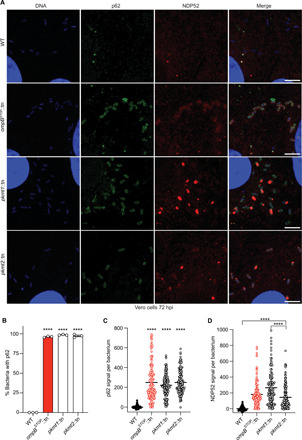
(A) Micrographs of Vero cells infected with the indicated strains at 72 hpi. Fixed with 4% PFA and stained for bacterial and host DNA (Hoechst; blue), p62 (anti-p62 antibody; green), and NDP52 (anti-NDP52 antibody; red) (representative of n = 4 for p62; n = 2 for NDP52). Scale bars, 5 μm. Image adjustments for each marker were applied equally for all the bacterial strains [except DNA (blue), which was adjusted slightly differently for WT bacteria]. (B) Percentage of bacteria that show rim-like surface localization with p62 at 72 hpi (n = 3). Statistical comparisons between all strains were performed using a one-way ANOVA with Tukey’s post hoc test. (C) Quantification of p62 signal per bacterium from a representative experiment (≥142 bacteria counted). Statistical comparisons were performed using a Kruskal-Wallis test with Dunn’s post hoc test. (D) Quantification and statistical comparisons of NDP52 signal per bacterium as in (C). ****P < 0.0001. Data are means ± SEM.
