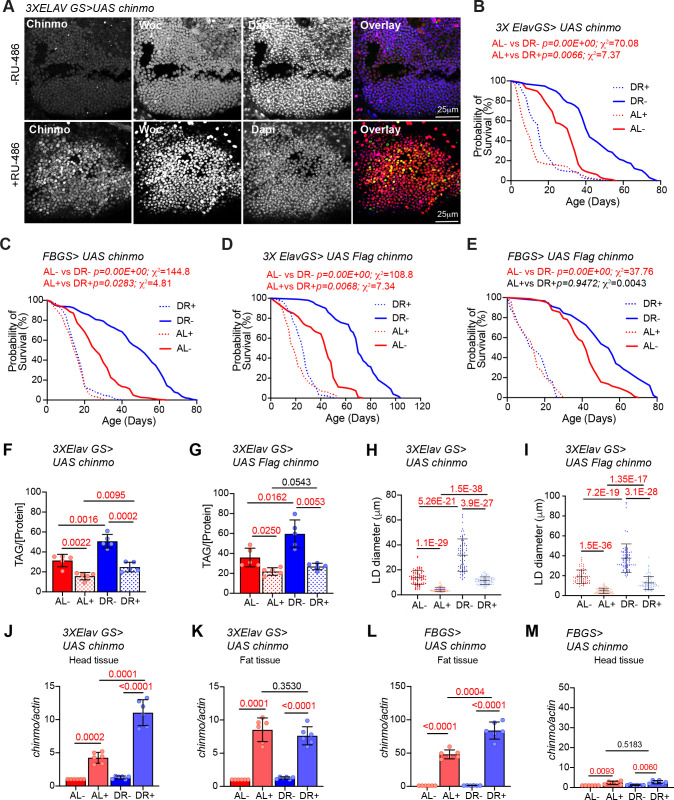
Figure 6. Ectopic expression of chinmo in adult neurons and fat body mimics miR-125 mutant DR phenotype.
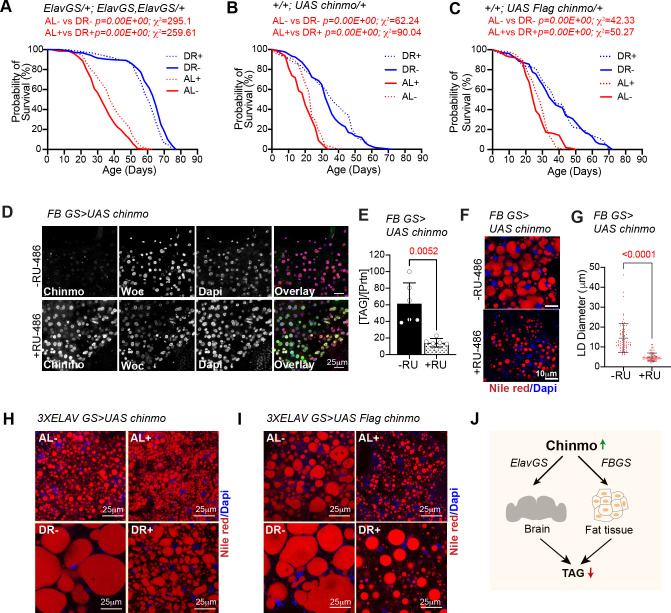
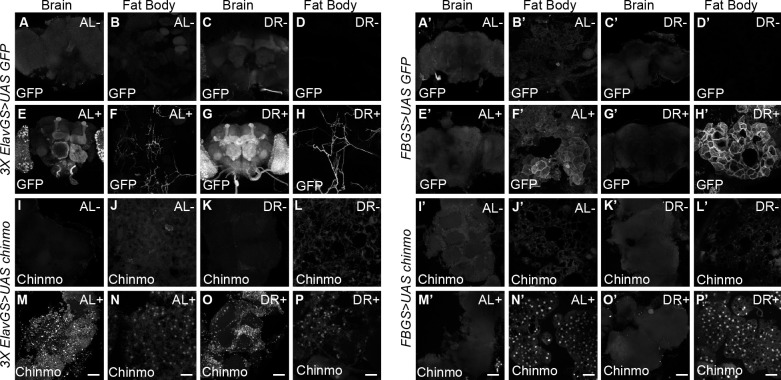
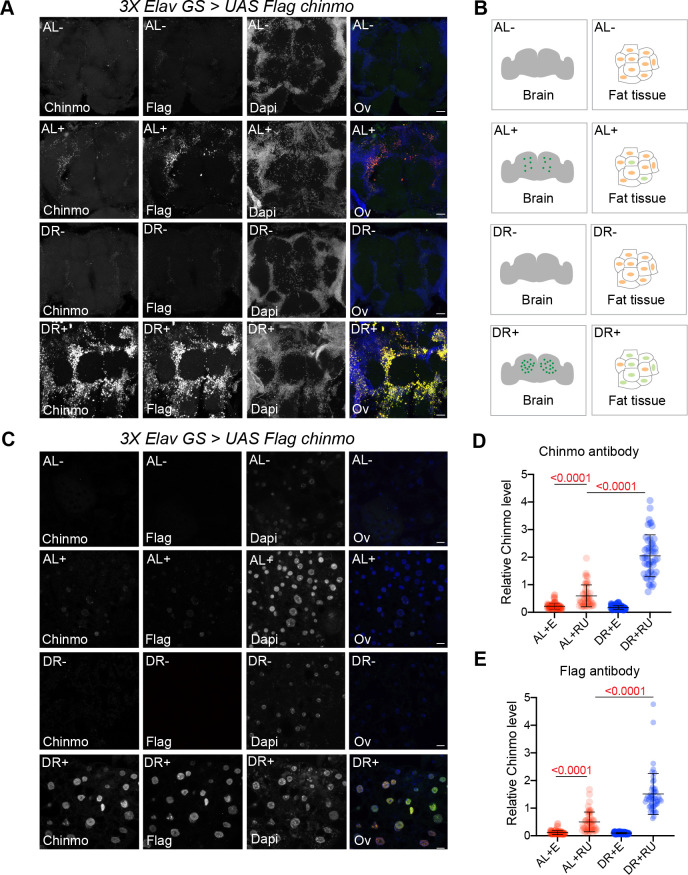
UAS chinmo was expressed in adult neurons and adult fat tissue using the steroid (RU-486) inducible gene switch Gal4 drivers. (A) Female flies that were fed an RU-486 supplemented diet for 5 days displayed increased levels of Chinmo in neuronal cells, as detected by Chinmo. Woc (nuclear marker) and Dapi staining of dissected adult fly brains. (B, D) 3X ElavGS >UAS chinmo (B) or 3X ElavGS >UAS Flag chinmo (D) flies that were not fed RU-486 show a significant increase in lifespan upon DR (DR, blue line) as compared to 3X ELavGS >UAS chinmo (B) or 3X ElavGS >UAS Flag chinmo (D) flies that were fed an ad libitum (AL) diet (red line)(3X ElavGS >UAS chinmo: AL-, n = 98; median lifespan = 32 d; DR-, n = 99; median lifespan = 42; 3X ElavGS >UAS Flag-chinmo: AL-, n = 107; median lifespan = 44 d; DR-, n = 99; median lifespan = 70 d). RU-486 fed 3X ElavGS >UAS chinmo flies or 3X ElavGS >UAS Flag-chinmo display a significantly dampened DR-dependent increase in lifespan (blue and red dotted lines) (3X ElavGS >UAS chinmo: AL+, n = 100; median lifespan = 10 d; DR+, n = 95; median lifespan = 16 d; 3X ElavGS >UAS Flag-chinmo: AL+, n = 137; median lifespan = 18 d; DR+, n = 100; median lifespan = 26 d). (C, E) FB GS >UAS chinmo or FB GS >UAS Flag-chinmo flies that were fed RU-486 displayed a stronger DR phenotype relative to 3X ElavGS >UAS Chinmo or 3X ElavGS >UAS Flag chinmo (blue and red dotted lines)(FB GS >UAS chinmo: AL-, n = 186; median lifespan = 26 d; DR-, n = 187; median lifespan = 52 d; AL+, n = 213; median lifespan = 16 d; DR+, n = 209; median lifespan = 16 d; FB GS >UAS Flag-chinmo: AL-, n = 96; median lifespan = 42; DR-, n = 91; median lifespan = 54; AL+, n = 83; median lifespan = 14 d; DR+, n = 73; median lifespan = 14 d) (Compare p values and Χ2 in panels B/D with C/E) compared to the flies that were fed the Ethanol (blue and red solid lines). For statistical comparison of survival curves, p values and Χ2 were calculated with log rank test. (F, G) Quantitation of triglyceride (TAG) stored levels in AL-RU-486 (Solid red bars), AL + RU-486 (Red dotted bars), DR-RU-486 (Solid blue bars) and DR +RU-486 (Dotted blue bars) fed 20d old (F) 3XElavGS > UAS chinmo or (G) 3XElavGS > UAS Flag-chinmo flies. Increasing Chinmo levels in adult neurons is sufficient to lead to a significant reduction in the systemic triglyceride levels under both AL and DR conditions. This decrease mimics the decrease in TAG levels observed in miR-125 mutants. The bars represent mean ± SD, n = 5, p value was calculated with unpaired two tailed t-test with Welch’s correction. (H, I) Fat bodies/abdomens of female flies were dissected and stained for the content and diameter of lipid droplet (LD) (red are lipid droplets stained with Nile red and blue is Dapi). For representative confocal images, please refer to Figure 6—figure supplement 1H–I. Quantitation of lipid droplet (LD) diameter in 3XElavGS > UAS chinmo (H) and 3XElavGS > UAS Flag-chinmo (I). A significant reduction is seen in lipid droplet size in the abdominal fat tissue when Chinmo is expressed in adult neurons. Quantitation of 15 largest LDs in five samples per condition. Error bars represent mean ± SD. (J–K) 3X ElavGS >UAS chinmo flies express increased levels of chinmo mRNA in both head and fat tissue. (L–M) FBGS >UAS chinmo flies predominantly express chinmo mRNA in the adult fat tissue. (J–M) Quantitative RT-PCR of RNA extracted from dissected tissues (head and fat body) of 3XElavGS > UAS chinmo (J, K) and FBGS >UAS chinmo (L, M) flies that were fed an AL (Red and pink bars) and DR (Dark Blue and Light blue bars) under uninduced (Red and Dark blue bars) and steroid induced (Pink and Light blue bar) conditions. Expression levels were normalized to Actin5c. Values are mean ± SD, n = 6. p Values were calculated using unpaired t test with Welch’s correction. Genotypes of strains used in this figure: (A, B, F, H, J-M) 3X ElavGS >UAS chinmo: P{elav-Switch.O}GS −1A / +; P{elav-Switch.O}GS-3A, P{elav-Switch.O} GSG301/P{w+, UAS-chin::SV40}/+; (D, G,I) 3X ElavGS >UAS Flag chinmo: P{elav-Switch.O}GS −1A / +; P{elav-Switch.O}GS-3A,P{elav-Switch.O} GSG301/P{w+, UAS Flag-chin::SV40} attP2 / +; (C) FBGS >UAS chinmo: w1118; P {w [+mW.hs]=Switch1}106/+; P{w+, UAS-chin::SV40}/+; (E) FBGS >UAS Flag-chinmo: w1118; P{w[+mW.hs]=Switch1} 106/+; P{w+, UAS Flag-chin::SV40} attP2 / +.