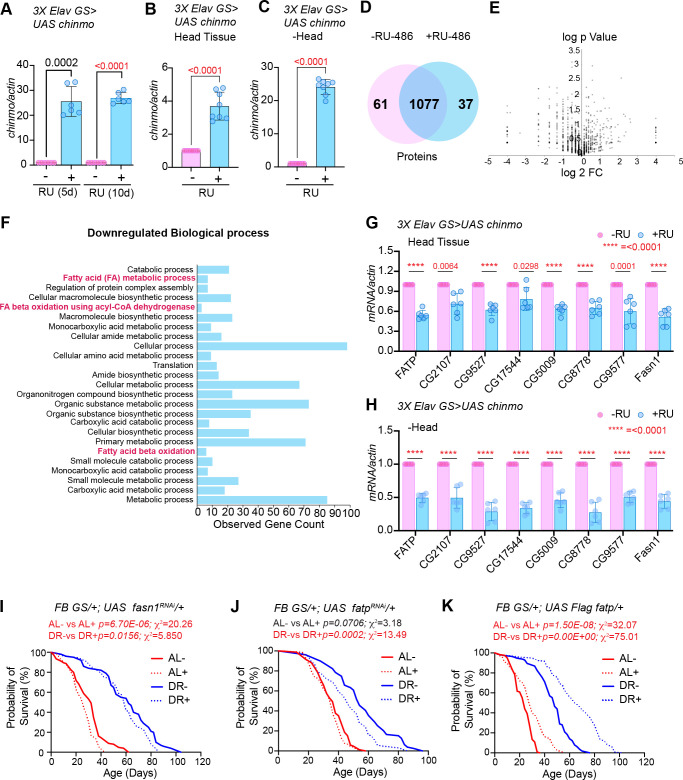
Figure 7. Ectopic expression of chinmo in adult neurons represses transcription of genes involved in fat metabolic processes.
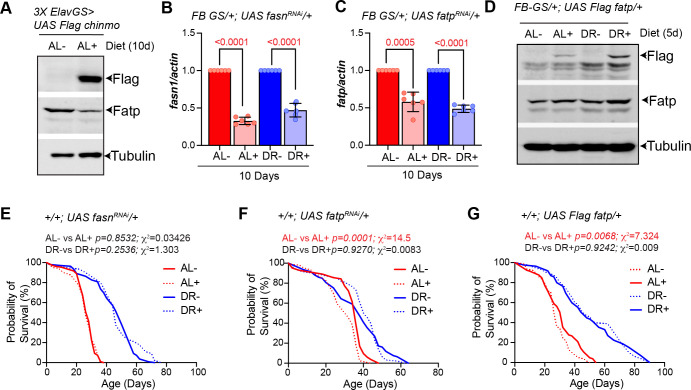
(A) Quantitation of chinmo mRNA levels in RNA extracted from 3X ElavGS >UAS chinmo flies that were fed a solvent (light blue bars) or RU-486 (pink bars) diet for 5 and 10 days. (B, C) Quantitation of chinmo mRNA levels in RNA extracted from head tissue (B) and decapitated body tissue (C) of 3X ElavGS >UAS chinmo flies that were fed ethanol (blue bar) or RU-486 (pink bar) diet for 10 days. Expression levels were normalized to Actin5c. Values are mean ± SD, n = 3. (D) Venn diagram of 1175 proteins identified across the two groups (-RU-486 and +RU-486) and 91% were common between the two groups. (E) Volcano plot illustrating significantly differentially abundant proteins. Forty proteins were found to be differentially expressed by using a cutoff on p value ≤ 0.05 and log2FC ≥ 1 (7 upregulated) and ≤−1.0 (33 downregulated) proteins. (F) Twenty-five most significant biological processes that are downregulated upon overexpression of chinmo in adult neurons. See Figure 7—figure supplement 2 for upregulated biological processes. (G, H) Overexpression of chinmo in the adult nervous system downregulates genes involved in fat metabolism. RT-PCR quantitation of fold- change in mRNA levels of genes (F) involved in fat metabolism in the head tissue (G) and decapitated body tissue (H) of 3X ElavGS >UAS chinmo flies that were fed the solvent/ethanol (light blue bars) or RU-486 (pink bars) diet for 10 days. (I) Knockdown of fasn1 in the adult fat body reduces lifespan more significantly in flies that are fed an AL (compare red solid and dotted lines) compared to flies that are fed a DR diet (compare blue solid and blue dotted lines) conditions (See Figure 7—source data 1 for p values and median and maximum lifespan for additional experimental replicate). (J) Knockdown of fatp in the adult fat body reduces lifespan under DR (compare blue solid and blue dotted lines) conditions (See Figure 7—source data 2 for p values and median and maximum lifespan for additional experimental replicate). (K) Overexpression of Flag FATP increases lifespan in flies that are fed a DR diet (Compare blue dotted line with blue solid line) more significantly than flies that are fed an AL diet (See Figure 7—source data 3 for p values and median and maximum lifespan for additional experimental replicate). For statistical comparison of survival curves, p values and χ2 were calculated with log-rank test. Genotypes of strains used in this figure: (A-H) 3X ElavGS >UAS chinmo: P{elav-Switch.O}GS −1A / +; P{elav-Switch.O}GS-3A, P{elav-Switch.O} GSG301/P{w+, UAS-chin::SV40}/+; (I) FB GS/+; UAS fasn1RNAi/+: w1118; P{w[+mW.hs]=Switch1}106/+; P{y[+t7.7] v[+t1.8]=TRiP.HMS01524}attP2/+; (J) FB GS/+; UAS fatpRNAi/+:w[1118]; P{w[+mW.hs]=Switch1}106/+; P{y[+t7.7] v[+t1.8]=TRiP.HMC04206}attP2/+; (K) FB GS/+; UAS Flag fatp/+: w[1118]; P{w[+mW.hs]=Switch1}106/+;P{w+, UAS-Flag fatp} attP2/+.
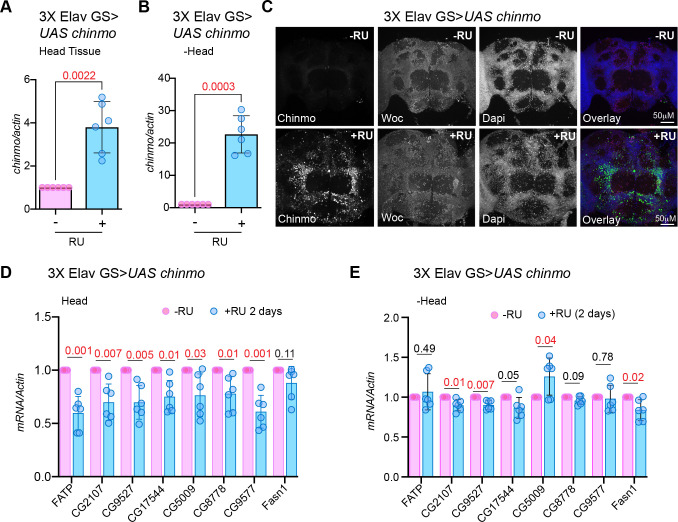
Figure 7—figure supplement 1. Overexpression of chinmo in adult neurons represses transcription of genes involved in fat metabolic processes in the brain within 2 days of induction.
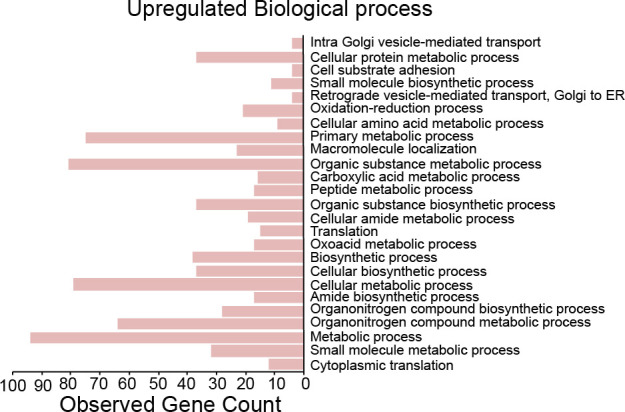
Figure 7—figure supplement 2. Over expression of chinmo in adult neurons upregulates cytoplasmic processes.