Abstract
KRAS, an oncogene, is frequently activated by mutations in many cancers. Kras-driven adenocarcinoma development in the lung, pancreas, and biliary tract has been extensively studied using gene targeting in mice. By taking the organoid- and allograft-based genetic approach to these organs, essentially the same results as in vivo models were obtained in terms of tumor development. To verify the applicability of this approach to other organs, we investigated whether the combination of Kras activation and Pten inactivation, which gives rise to endometrial tumors in mice, could transform murine endometrial organoids in the subcutis of immunodeficient mice. We found that in KrasG12D-expressing endometrial organoids, Pten knockdown did not confer tumorigenicity, but Cdkn2a knockdown or Trp53 deletion led to the development of carcinosarcoma (CS), a rare, aggressive tumor comprising both carcinoma and sarcoma. Although they originated from epithelial cells, some CS cells expressed both epithelial and mesenchymal markers. Upon inoculation in immunodeficient mice, tumor-derived round organoids developed carcinoma or CS, whereas spindle-shaped organoids formed monophasic sarcoma only, suggesting an irreversible epithelial-mesenchymal transition during the transformation of endometrial cells and progression. As commonly observed in mutant Kras-driven tumors, the deletion of the wild-type Kras allele was identified in most induced tumors, whereas some epithelial cells in CS-derived organoids were unexpectedly negative for KrasG12D. Collectively, we showed that the oncogenic potential of KrasG12D and the histological features of derived tumors are context-dependent and varies according to the organ type and experimental settings. Our findings provide novel insights into the mechanisms underlying tissue-specific Kras-driven tumorigenesis.
Subject terms: Endometrial cancer, Cancer stem cells
Introduction
Ras is a small guanosine-5’-triphosphate (GTP)-binding protein that transmits external stimuli to downstream signaling pathways, such as Raf, PI3K, and Ral-GEF. These effectors orchestrate various vital cellular processes, including gene transcription, cell proliferation, acquisition of cell motility, and inhibition of cell death [1]. Although Ras activation is normally regulated through GTP-binding and degradation in a transient and reversible manner, its constitutive activation is often observed in various types of cancer, mostly by missense mutations in the GTP-binding sites at codons 12, 13, and 61 or by gene amplification [2]. Epithelial malignancies have preferences for KRAS mutations among the three RAS genes, namely HRAS, NRAS, and KRAS; although KRAS mutation rate varies among tissues, it can be as high as ~90% in pancreatic cancer; 20–50% in carcinomas of the colon, lung, and hepatobiliary tract; and much less frequent in other carcinomas [3]. Using genetically engineered mice (GEM) models, many studies have demonstrated the causal role of the KrasG12D mutation in tumor development, through cooperation with other common genetic aberrations in each cancer type [4]. Thus, KrasG12D has been established as a bona fide oncogene.
Endometrial cancer (EC) is the most common gynecological malignancy and the fourth most common female cancer in industrial countries [5,6,]. EC is typically classified into two types based on clinical and histological characteristics. Type 1, comprising 80% of all ECs, is associated with estrogen dependency, endometrial hyperplasia, and a favorable prognosis. The predominant histological subtype is endometrioid carcinoma, harboring frequent mutations in PTEN (52–78%), PIK3CA (36–52%), KRAS (15–43%), ARID1A (25–48%), and CTNNB1 (23–24%). In contrast, type 2 is associated with estrogen independency, endometrial atrophy, and poor prognosis, and comprises various minor subtypes including serous carcinoma, clear cell carcinoma, and carcinosarcoma (CS), with high mutation rates in TP53 (60–91%) [7,8,]. Not in a mutually exclusive manner with these recurrent mutations, microsatellite instability (MSI) is found in 40% of endometrioid carsinoma and 2% in serous carcinoma, thereby leading to the proposal of a novel classification based on the mutational burden; the four categories included POLE-mutated (ultramutated), MSI (hypermutated), copy-number low, and copy-number high, the first two of which have significantly better prognosis [7].
Because PTEN has the highest mutation rate across EC, the effect of its inactivation has been extensively investigated using GEM. Although Pten−/− mice died during embryogenesis [9], all Pten+/− mice developed precancerous lesions, 25% of which eventually developed EC within a year [10]. PR-Cre; Ptenflox/flox mice, in which Pten was deleted by the Cre-recombinase induced in a uterine-specific manner, frequently developed invasive EC by 3 months of age [11]. After the local injection of adenovirus Cre into the uterus of Ptenflox/flox mice, EC development was observed, albeit with partial penetrance [12,13,]. Concurrent KrasG12D accelerated Pten-dependent EC development [14,15,], suggesting that KrasG12D plays an oncogenic role in the endometrium. Alternatively, GEM with genitourinary tract-selective Trp53 deletion developed type 2 EC in 84% of mice at 58–68 weeks of age [16]. These findings suggest that the tumorigenic potential of endometrial cells and the histological features of EC may be genetically determined to a considerable extent, although more studies are required to fully elucidate its pathogenesis.
Organoid culture is an emerging technology that enables the long-term propagation of normal epithelial cells in a physiological setting [17,18,]. It is a three-dimensional culture method using the Matrigel, extracellular matrix that mimics the basement membrane, and serum-free medium supplemented with stem cell niche factors for each tissue. Owing to the high utility, its application has been expanding to many research fields, including infectious diseases [19], developmental biology [20], and tissue regeneration [21]. We reconstituted a combination of common genetic aberrations in organoids with lentiviral vectors encoding short hairpin RNA (shRNA) and Cre, which were inoculated into nude mice. Even without a tissue-specific microenvironment, certain combinations of genetic aberrations that are particularly common in certain cancers robustly give rise to subcutaneous tumors. For example, Apc inactivation and Kras activation in the intestinal organoids markedly accelerated tumorigenesis caused by Apc knockdown alone [22]. In organoids from the lung [23], hepatobiliary tract [24,25,], and pancreas [26], the development of full-blown tumors was achieved not by Kras activation alone, but by the concurrent inactivation of the p53 or Rb pathway. Since these outcomes are consistent with the results of earlier GEM studies [22,23,25,26,], we hypothesized that such organoid and allograft-based approach can establish an alternative carcinogenesis model in any organ.
To verify this notion, we investigated whether organoids from the murine endometrium could be transformed by reconstitution of genetic aberrations that developed type 1 EC in GEM. Through examination of transduced organoids, subcutaneous tumors, and derived organoids, we gained novel insights into multi-layered interactions that are involved in tumor development.
Results
Pten knockdown allowed endometrial organoids to propagate after lentiviral infection
To conduct an organoid-based tumorigenicity assay, we isolated and propagated primary epithelial cells from the murine uterine horn (Fig. 1A). Endometrial organoids that robustly propagated for at least a few months had rounded cystic structures, whereas stromal cells spontaneously disappeared within a few passages (Fig. 1B). Organoids consisted of columnar epithelial cells lined in a monolayer as verified by the cytokeratin+/vimentin- staining pattern (Fig. 1C). Efficient gene transduction was verified using a lentivirus vector encoding GFP (Fig. 1D).
Fig. 1. Endometrial organoids with Pten knockdown exhibited low tumorigenic potential.
A Macroscopic view of the gynecologic tract. A portion of the uterine horn (red rectangle) was dissected. Scale bar, 2.5 mm. B Representative images of endometrial organoids. Time-lapse images of primary (P0) cells and after 3 passages (P3) are shown. Scale bar, 200 μm. C Immunohistochemical (IHC) staining. Upper panel, organoids. Lower panel, uterine tissue. Immunostaining results of serial sections with antibodies against cytokeratin or vimentin are shown. Scale bar, 30 μm. D Lentiviral gene transduction of endometrial organoids. The same views of GFP-transduced organoids are shown. Scale bar, 200 μm. E Western blot analysis of transduced organoids. Both Pten knockdown and Akt phosphorylation were achieved by shPten. The induction of p16Ink4a is prominent following lentiviral transduction with shLuc, but not with shPten. F Macroscopic findings of an isolated nodule. Pten knockdown alone did not result in the tumorigenic potential of endometrial organoids. Scale bar, 10 mm. G Histological findings of nodules derived from organoids with Pten knockdown. A few glands with mild atypia are observed after H&E staining. Scale bar, 50 μm.
Considering that most previous GEM models for EC were generated by the ablation of Pten [9–13], we first evaluated the tumorigenic potential of the organoids with Pten knockdown. By introducing shRNAs against Pten (hereafter, shPten) into the endometrial organoids, Pten knockdown and the subsequent Akt activation were verified compared to the shRNA against luciferase (shLuc) (Fig. 1E). Although organoids without infection or with shPten continued to proliferate, those with shLuc failed to propagate after the first passage and had a considerable elevation of the cell cycle inhibitor p16Ink4a, suggesting the deleterious effects of lentiviral infection. Upon the inoculation of organoids with shPten into nude mice, only tiny nodules (Fig. 1F) containing a few glands with mild atypia of ductal or cystic shape developed (Fig. 1G). These observations suggest that despite the advantages of in vitro proliferation and unlike earlier in vivo studies, Pten knockdown alone is insufficient to induce tumorigenesis in endometrial organoids.
Endometrial organoids with KrasG12D and Pten knockdown did not develop tumors
The activation of the Ras pathway has been implicated in human EC [7]. As KrasG12D accelerated the Pten-dependent EC development in mice [14,15,], we investigated whether this combination of genetic alterations could drive tumor development in endometrial organoids. We applied a two-step gene transduction protocol, namely, introduction of Cre and shRNA transduction (Fig. 2A). Organoids from KrasLSL-G12D/+ mice were subjected to lentiviral infection, in which KrasG12D is transcribed upon the Cre-mediated excision of the STOP codon flanked by the two LoxP sequences (LSL) [28]. Subsequently, we detected a newly emerged KrasG12D amplicon slightly longer than the KrasWT amplicon by a single LoxP (Fig. 2B) and observed the induction of the KrasG12D protein (Fig. 2C), establishing successful recombination.
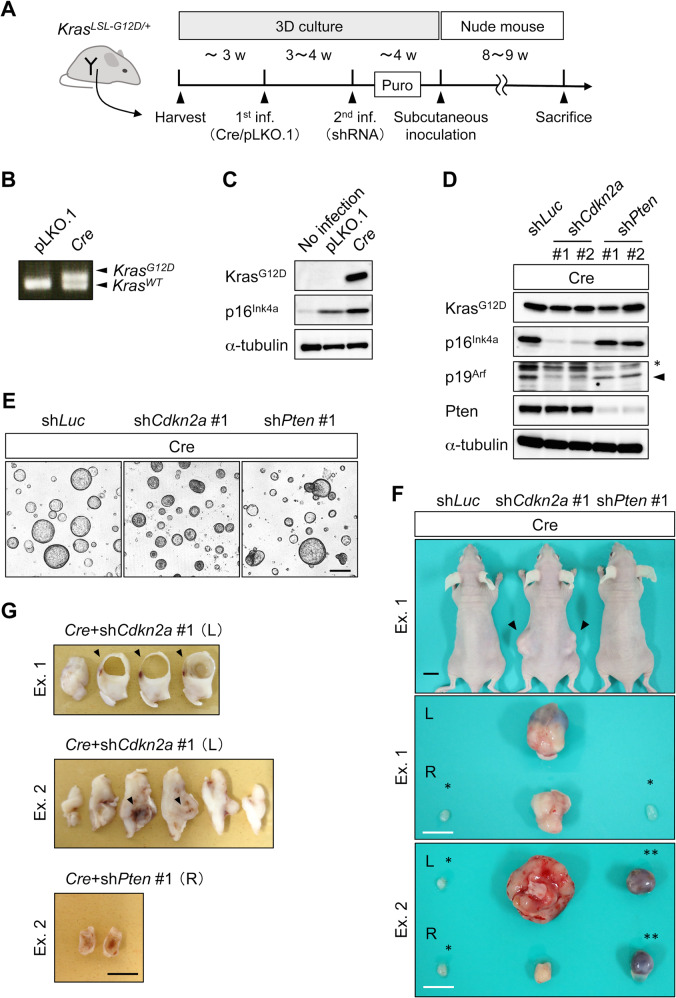
Fig. 2. KrasG12D organoids with Cdkn2a knockdown developed CS.
A Two-step protocol for organoid generation from KrasLSL-G12D/+ mouse. B Genomic PCR analysis of transduced organoids. The upper band depicted as KrasG12D indicates the successful recombination of the Kras locus by Cre-recombinase, whereas the lower band depicts an amplicon for the WT allele of Kras. C Western blotting analysis of endometrial organoids. Stepwise inductions of p16Ink4a by infection itself and Cre transduction were observed. D Western blot analysis of Kras-activated endometrial organoids. The knockdown of the targeted gene products was achieved by the introduction shCdkn2a and shPten clones. shCdkn2a suppressed its two products p16Ink4a and p19Arf. Non-specific bands (*) and specific bands (arrowhead) are shown for the p19Arf panel. E Transduced organoids visualized under phase-contrast microscope. Scale bar, 200 μm. F Tumor development in nude mice. Upper panel, subcutaneous tumors (arrowheads). Scale bar, 10 mm. Middle and lower panels, resected tumors. Representative results from two independent experiments (Exs. 1 and 2) are shown. Matrigel plugs (single asterisk) and cystic lesions (double asterisks) are classified as non-tumorous lesions. Scale bar, 10 mm. G Macroscopic view of sliced tumors after formalin fixation. Scale bar, 10 mm. Cre + shCdkn2a #1: a solid tumor with cystic formation (arrowhead) in Ex. 1 and with necrosis (arrowhead) in Ex. 2. Cre + shPten #1: multilocular cyst in Ex. 2.
Although organoids with Cre steadily proliferated, those with the backbone vector pLKO.1 gradually stopped propagating over passages, similar to the organoids after transduction with shLuc. Because p16Ink4a was induced after lentiviral infection and more prominently after KrasG12D induction, two potent shRNA clones against Cdkn2a encoding p16Ink4a and p19Arf were added to the second infection, along with shPten and shLuc, to avoid cell cycle arrest (Fig. 2D). No morphological changes were observed (Fig. 2E). Most KrasG12D organoids with shLuc did not develop tumors in the nine cases tested (Fig. 2F), with the exception of one case of CS, which was defined by the presence of both carcinoma and sarcoma components in the tumor (Fig. S1, Table S1). KrasG12D organoids with shPten clone #1 developed cysts in two out of three cases (Fig. 2F, G), in which the lumen was covered with a monolayer of columnar cells with mild atypia (Fig. 3A, B). Another shPten clone #2 did not induce tumors in two experiments, even after introduction into the KrasG12D organoids that gave rise to sarcoma upon the introduction of shLuc (Table S1, Fig. S1). These observations suggested the marginal tumorigenic potential of KrasG12D, which was not promoted by Pten knockdown.
Fig. 3. Histological features of organoid-derived CS and monophasic sarcoma.
(A) H&E staining of thinly sliced sections. Cre + shCdkn2a #1 is a sarcoma with cyst (arrowheads), whereas Cre + shPten #2 is a multilocular cyst. Scale bar, 10 mm. (B) H&E staining of thinly sliced sections of subcutaneous nodules from Exs. 1 and 2. Matrigel plugs induced by Cre + shLuc contained only a few glands with mild atypia. Tumors induced by Cre + shCdkn2a #1 are sarcoma in Ex. 1 and carcinosarcoma in Ex. 2. Matrigel plugs (Ex. 1) and that by Cre + shPten #1 is a cystic lesion (Ex. 2). Scale bar, 50 μm. (C) Characterization of heterogeneous lesions of carcinosarcoma. H&E and IHC staining against cytokeratin and vimentin of serial sections for areas 1–3 are shown. Magnified view of the corresponding areas is shown. Dotted lines in area 1 indicate cytokeratin+/vimentin-. Scale bar, 50 μm. (D) H&E staining of the tumors induced by Cre + shCdkn2a #2. Cystic lesion from Ex. 1 and sarcoma lesion from Ex. 2 are shown. Scale bar, 50 μm.
KrasG12D organoids with Cdkn2a knockdown or Trp53 deletion developed CS
In contrast, KrasG12D organoids with shCdkn2a developed solid tumors (Fig. 2F) within eight weeks in all five cases (Table S1). The tumors partly contained cystic lesions or necrosis (Figs. 2G, 3A) and were predominantly occupied by spindle-like cells and partly by atypical glandular cells. Tiny nodules originating from the KrasG12D organoids with shLuc only contained a few glands with mild atypia (Fig. 3B). Among the five solid tumors, two were diagnosed with CS, two with monophasic sarcoma, and one with a combination of sarcoma and cyst (Table S1). In the CS cases, most carcinoma and sarcoma cells separately resided as cytokeratin+/vimentin- and cytokeratin-/vimentin+ populations, respectively (Fig. 3C). However, a subset of sarcoma cells showed cytokeratin+/vimentin+ expression, indicating an intermediate state between typical carcinomas and sarcomas (Fig. 3C). This finding suggested an epithelial origin of the sarcoma cells. In the two experiments with another potent shCdkn2a clone, cysts or sarcomas were induced in KrasG12D organoids (Fig. 3D), thus eliminating the possibility of off-target effects by shRNA. These results suggest that KrasG12D expression and Cdkn2a knockdown cooperate for tumor development, preferentially toward the induction of sarcomatous differentiation.
In human uterine CS, TP53 is the most frequently mutated gene [29,30,]. Consistent with this finding, GEM with uterine-specific Trp53 deletion developed CS after a long latency and only in ~10% of the tumors [16]. This suggested that other genetic alterations were required to accelerate CS development; therefore, we investigated whether KrasG12D and p53 loss could synergize tumorigenesis. After Cre transduction of endometrial organoids from KrasLSL-G12D/+; Trp53flox/flox mice (Fig. 4A), the simultaneous induction of KrasG12D expression and Trp53 deletion (Figs. 4B, 4C) was verified. We observed the steady proliferation of organoids (Fig. 4D), but not in organoids with pLKO.1 as predicted. Upon the inoculation into nude mice, solid tumors developed within eight weeks in all four cases (Fig. 4E). Notably, the tumors were invariably diagnosed as CS (Table S1, Fig. 4F). Thus, Trp53 deletion also cooperated with KrasG12D expression for CS development in endometrial organoids.
Fig. 4. KrasG12D organoids with deleted Trp53 developed CS.
A One-step protocol for organoid generation from KrasLSL-G12D/+; Trp53flox/flox mice. B Genomic PCR analysis of endometrial organoids from KrasLSL-G12D/+; Trp53flox/flox mice. The presence of the PCR products for the amplicons of KrasG12D and the recombined allele of Trp53 indicates the successful Cre-mediated recombination in both loci. C Western blot analysis of transduced organoids. Induction of KrasG12D and loss of p53 at the protein level were demonstrated. D A representative image of endometrial organoids after Kras activation and Trp53 deletion. Images for mock-infected organoids are not shown owing to the lack of steady propagation after lentiviral infection. Scale bar, 200 μm. E Tumor development following Kras activation and Trp53 deletion. Upper panel, a representative image of subcutaneous tumors developed in nude mice. Lower panels, resected tumors. Representative results of two independent experiments (Exs. 1 and 2) are shown. Scale bar, 10 mm. F Histological findings in subcutaneous tumor. Upper panel, H&E staining of thin sections for the same four slices illustrating multiple cystic structures in the solid tumor (asterisks). Lower panel, Magnified images of the two areas (boxes) are shown. Note that the area 1 consists exclusively of spindle-like, pleomorphic cells, whereas area 2 contains carcinoma cells showing a glandular structure. Scale bar, 50 μm.
Tumor-derived organoids retained epithelial-mesenchymal transition (EMT) state
Regardless of the presence of genetic alterations, endometrial organoids stereotypically maintained their round cystic shape. In contrast, those recovered from subcutaneous CS or sarcoma displayed diverse morphological features ranging from cystic, spindle-like, and combined types (Fig. 5A). Previously, we revealed that nude mice-derived stromal cells could not survive the current serum-free culture conditions over several passages [25,26,]. The presence of spindle-like cells in six out of eleven tumor-derived organoids (TDOs) strongly suggests that their transformed nature was stably retained during recovery from subcutaneous tumors. In addition, one combined type and two spindle-like type TDOs developed CS and monophasic sarcoma, respectively, whereas two cystic-type TDOs developed monophasic carcinoma or CS (Fig. 5A, Table S2). These observations indicated that spindle-like cells might have originated from epithelial cells and that carcinoma cells could have undergone EMT to sarcoma in an irreversible manner.
Fig. 5. Morphological and genomic heterogeneity among tumor-derived organoids.
A Morphological classification of organoids recovered from subcutaneous tumors. Upper panel, mixed type (#1) contained both spindle-like cells that protrude from the organoids (white arrowhead) and standard cystic organoids (black arrowhead). Cystic type (#5 and #6) contained only standard cystic organoids, whereas spindle-type (#8) contained only spindle-like cells with occasional cell aggregation (red arrow heads). Scale bar, 200 μm. Lower panel, H&E staining of tumors induced by the re-inoculation of subcutaneous tumor-derived organoids. Mixed type induced the formation of carcinosarcoma (CS) (#1); cystic type, adenocarcinoma (#5), or CS (#6); and spindle-type sarcoma (#8). Scale bar, 50 μm. B Genomic PCR analysis of the Kras locus in tumor-derived organoids. C Copy number alterations in transduced or tumor-derived organoids as identified by array CGH analysis. SCT, subcutaneous tumor; LucKD, introducing shLuc.
Tumor-derived organoids displayed heterogeneous statuses in the Kras loci
For pancreatic and biliary tract organoids, TDOs generated from organoid-based Kras-driven carcinogenesis models were completely negative for the LSL cassette in the conditional allele [25,26,]. Considering that a fraction of cells retained the LSL at the time of inoculation, this finding suggested that Kras activation was a definite requirement for adenocarcinoma development in these organoids. Similarly, we examined the Kras locus in nine endometrial TDOs using genomic PCR analysis (Fig. 5B, Table S2). The emergence of the KrasG12D amplicon was detected in all TDOs as predicted, whereas the residual LSL cassette was unexpectedly detected in some cases (Fig. 5B, Table S2), indicating the survival of untransformed cells without KrasG12D within the CS. In addition, the intensity of KrasWT amplicon was clearly fainter than that of KrasG12D in four cases and was undetectable in five cases (Fig. 5B, Table S2), suggesting the spontaneous deletion of the KrasWT allele, as occasionally observed in the organoid-based pancreatic cancer model [26].
To evaluate the genetic instability comprehensively during CS development, we performed array CGH analysis on three pairs of transduced organoids and their corresponding TDOs. For KrasG12D organoids with both Cdkn2a knockdown (#4) and Trp53 deletion (#5, #6), the pre-inoculated organoids and TDOs exhibited genome stability, although no recurrent amplifications or deletions were detected (Fig. 5C). Thus, the Kras loci may have undergone unique alterations during the development of CS and sarcoma.
Significant transcriptomal changes in organoids underlay tumorigenesis
To better characterize the pathogenesis of CS from organoids, we conducted a transcriptome analysis. Based on hierarchical cluster analysis, organoids before inoculation and TDOs (#5 and #6) were relatively similar, although a spindle-type TDO (#4) was observed as an outlier (Fig. 6A). The Kyoto Encyclopedia of Genes and Genomes database was used to identify signaling pathways that were significantly altered during tumorigenesis. Among the up-regulated gene functions, three out of the top ten were shared by all TDOs, including ECM-receptor interaction, the PI3K-AKT signaling pathway, and cytokine-cytokine receptor interaction (Fig. 6B). Considering that CS and sarcoma likely originated from epithelial organoids, we further examined the expression of EMT marker genes in TDOs. Spindle-type organoids (#4) showed high expression levels of several known mesenchymal markers, such as Vimentin, Cdh2, Zeb1, and Snai1, and low expression levels of epithelial markers, such as Krt7, Cdh1, Cldn7, and Ersp1 [31] (Fig. 6C). In contrast, the transcriptome profile of cystic-type organoids (#5 and #6) was similar to that of pre-inoculated organoids, except for a moderate increase in Vimentin (Fig. 6C). These results indicate that gene expression profiles reflect the morphology of TDOs.
Fig. 6. Transcriptomic changes in organoids after inoculation in nude mice.
A Heat map showing differentially expressed genes in transduced or tumor-derived organoids as identified by using microarray analysis. Colors range from blue to red corresponding to genes with low and high expression, respectively. B Activated function was inferred from up-regulated genes in each tumor-derived organoid. C Gene expression of epithelial–mesenchymal transition markers in tumor-derived organoids.
Discussion
We previously showed high concordance of tumorigenicity caused by certain combinations of genetic aberrations between GEM and organoid-based carcinogenesis models [22,25,26,]. Regarding Pten-driven tumorigenicity in the uterus, tumor development was not demonstrated in this study, even in the presence of KrasG12D. One possible explanation for the observed discrepancy is that the uterine-specific gene targeting in mice by PR-Cre mice [11] or the local injection of adeno-Cre [12,13,] may in fact have resulted in gene recombination in both epithelial and stromal cells of the uterus. In line with this notion, mice with uterine epithelial-specific ablation of Pten did not develop EC [32]. Considering that the resultant Pten knockdown in the stroma is not common in sporadic cases of human EC, the overestimated tumorigenic potential could be attributed to the interplay between the epithelia and the stroma of the uterus. Similar observations were previously documented in intestinal tumorigenesis, wherein Pten deletion in both the stroma and epithelia in mice led to tumor development, which was not observed when deletion was specific to the epithelium [33,34,] Another possibility is that even normal uterine stroma could play a pro-tumorigenic role. In a cell-based study, the introduction of myr-Akt or shPten into primary murine endometrial cells, followed by co-transplantation with the uterine stroma from neonatal WT mice, resulted in the development of adenocarcinoma in the kidney capsule [35], suggesting the importance of the microenvironment of the uterine stroma in EC development. Lastly, given that the MSI subtype of human EC preferentially harbors mutations in KRAS and PTEN [7], it is tempting to speculate that additional inactivation of mismatch repair genes in endometrial organoids could achieve type I EC development, which is worthy of further investigations.
Another unique property of endometrial organoids is the propagation arrest associated with lentiviral infection, which has never been observed in any gastroenterological organoids under almost the same culture conditions [22–26]. Considering the infinite propagation of uninfected organoids and p16Ink4a induction following lentiviral infection, endometrial organoids may be particularly sensitive to DNA damage caused by viral genomic integration. Although the underlying mechanisms remain elusive, this finding prompted us to knockdown Cdkn2a in endometrial organoids, which unexpectedly led to the Kras-dependent development of CS or sarcoma. In human EC cases, KRAS mutations are rarely found in serous carcinoma (3%) but are more prevalent in endometrioid carcinoma (24%) and CS (12%); TP53 mutations are significantly more common in serous carcinoma (88%) and CS (91%) than in endometrioid carcinoma (21%) [36]. These findings are consistent with CS development in endometrial organoids upon Kras activation and p53 loss in this study. In GEM with identical genetic alterations, CS development was also observed in the ovary [37], while adenocarcinoma was always induced in the pancreatobiliary system for both in vivo and ex vivo [25,26,38,39,]. These findings suggest that gynecological organs may be predisposed to CS due to some inherent epigenetic regulation.
We further demonstrated the development of CS from epithelial cells in two ways: from transduced cystic organoids and from cystic-type TDOs originating from organoid-derived CS. In addition, we identified a fraction of CS cells that expressed both epithelial and mesenchymal markers. These findings are consistent with the notion that uterine CS is predominantly a monoclonal neoplasm of epithelial origin, rather than generated by the collision of two distinct cell populations [40,41,]. This indicates irreversible EMT during transformation and progression from carcinoma to sarcoma. Single-cell analysis at each step of this model may help clarify the molecular events involved in CS development. Further investigation on whether our results can be extrapolated to in vivo GEM is warranted. Also, our study might provide a unique resource for CS research, in which models are still few.
CS induced in this study was “homologous”, which by definition demonstrates the presence of spindle-like sarcoma cells resembling inherent uterine stroma, such as fibroblasts and smooth muscle cells. In contrast, the uterine epithelial-specific ablation of Fbxw7 and Pten in mice resulted in the development of “heterologous” CS, having a sarcoma component of differentiation toward bone and cartilage in half of the cohorts in three months [42]. Interestingly, mutations in Trp53 and Kras were found in four and one cases, respectively, among 76 late-stage heterologous CS cases, suggesting the relevance of the deregulation in these two genes in the common pathway to the development of uterine CS. In line with this notion, tumors with concurrent mutation in KRAS and TP53 distributed across all four subtypes of uterine CS, which respectively correspond to the same four categories in EC [43]. We speculate that certain genetic alterations may determine the differentiation of cell lineages and molecular subtypes of CS, although further investigation is required.
The characterization of the Kras locus in organoids and TDOs revealed several aspects of CS pathogenesis. First, the copy number of the KrasWT allele frequently decreased in TDOs. We have previously documented the complete loss of the KrasWT allele, albeit at a low frequency, in KrasG12D-expressing TDOs originating from pancreatic organoids [26]. The frequent loss of KRASWT has also been reported in tumors with mutant KRAS in humans [44]. Collectively, these findings are consistent with the notion that KrasWT acts as a relative tumor-suppressor gene in the presence of oncogenic Kras and that its deletion leads to the hyperactivation of the Kras pathway [45]. Second, the complete loss of KrasWT was observed in the TDOs. Because this is unlikely to occur spontaneously in normal cells, this observation negates the possibility that host-derived stromal cells can transform into CS cells. Last, the presence of residual cells without recombination in the Kras locus was revealed in TDOs. This finding is contrary to previous observations that TDOs originating from adenocarcinoma comprises only cells expressing KrasG12D, but not those that retain KrasWT, thus highlighting the critical roles of Kras activation in the development of adenocarcinoma [25,26,]. Consequently, based on this unexpected finding, we speculated that uterine CS cells might support the survival of normal epithelial cells to retain KrasWT, which might subsequently promote the proliferation of CS cells.
In conclusion, we established uterine CS using an organoid-based approach. Although patient-derived organoids (PDOs) for normal and cancer tissues in gynecologic organs have been recently established [27, 46–49], to the best of our knowledge, there are almost no reports on PDOs from CS cases. Therefore, our generated tumor organoids will be useful in preclinical studies of CS. In addition, future comparisons among organoids derived from ex vivo carcinogenesis models, GEM, and PDOs can provide further insights into the mechanisms underlying endometrial tumorigenesis.
Materials and methods
Mice studies
Female mice of the C57BL/6 J strain and Balb/cAnu/nu (nude mice) at 5 weeks of age were purchased from CLEA Japan Inc. (Tokyo, Japan). Conditional knock-in mice heterozygous for the Lox-STOP-Lox-KrasG12D allele (hereafter referred to as KrasLSL-G12D/+ mice) and Trp53flox/flox mice were obtained from the Jackson Laboratory (Bar Harbor, ME, USA) and maintained in a C57BL/6 J background. These mice were intercrossed to generate KrasLSL-G12D/+; Trp53flox/flox mice. Genotyping was performed after weaning as previously described [28,50,]. Animal studies were carried out with the approval of the Chiba Cancer Center for Ethics in Animal Experimentation.
Isolation and organoid culture of endometrial cells
Bilateral uterine horns were isolated from C57BL/6 J mice at approximately 10 weeks of age. The tissues were minced into 2–3 mm pieces, washed several times with cold PBS, and dissociated with 2 U/mL dispase II and 1 mg/mL collagenase P (Roche Diagnostics K.K., Tokyo, Japan) for 30 min at 37 °C. Dissociated cells were subjected to the Matrigel bilayer organoid culture (MBOC) protocol [22]. Briefly, cells were resuspended in the medium supplemented with L-glutamine solution (Wako, Osaka, Japan), penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO), amphotericin B suspension (Wako), 50 ng/mL EGF (Peprotech, Rocky Hill, NJ), 250 ng/mL R-spondin1 (R&D, Minneapolis, MN), 100 ng/mL Noggin (Peprotech), 10 μM Y27632 (Wako, Osaka, Japan), 1 μM Jagged-1 (AnaSpec, Fremont, CA), and 2.5 μM CHIR-99021 (Cayman Chemical, Ann Arbor, MI), and plated on 65 μL of solidified Matrigel (BD Biosciences, Franklin Lakes, NJ) per well in a 12-well plate. After overnight incubation at 37 °C, floating or dead cells were removed, leaving only viable cells attached onto Matrigel, which were covered with 70 μL of Matrigel and the medium. Passage was conducted every 5–10 days at 1:2 to 1:3 dilution. In each passage, organoids were dissociated using Accutase (Innovative Cell Technologies, San Diego, CA) for 5 min at 37 °C and vigorous pipetting, followed by seeding on Matrigel.
Lentiviral vectors and infection
Lentiviral particles were generated using the ViraPower Lentiviral Expression System (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. The collected viral supernatants were passed through a 0.45-µm filter, concentrated 10-fold with PEG-it Virus Precipitation Solution (System Biosciences), and stored at −80 °C until further use. The lentiviral transduction of organoids was performed as previously described [51]. Puromycin selection (3 µg/mL) was conducted for 4–5 days. For shRNA transduction, pLKO.1-puro vectors (Sigma-Aldrich, St. Louis, MO) targeting murine Cdkn2a (TRCN222731 and TRCN231227) and Pten (TRCN322421 and TRCN28992) were used. As a negative control shRNA, shLuciferase (SHC007) was used. LV-Cre pLKO.1 (Addgene plasmid 25997) [52] was used to remove the sequence flanked by the two LoxP sequences. Cre-mediated recombination was verified by genomic PCR as previously described [28,50,]. pCDH-CMV-MCS-EF1-copGFP (System Biosciences, Mountain View, CA) was used to monitor transduction efficiency.
Tumorigenicity assays in nude mice
Transduced endometrial cells were propagated as organoids in Matrigel. Organoids corresponding to 5 × 105 cells were resuspended in 200 μL of medium mixed with Matrigel at a 1:1 ratio and inoculated into the dorsal skin of nude mice. After 8–9 weeks, palpable tumors or residual Matrigel plugs from the injected sites were isolated for histological examination or cell culture. The tumors were processed essentially in the same way as cell preparation for the primary organoid culture. As serum-free culture media do not support the survival of tumor-derived stromal cells, a pure population of epithelial cells is normally obtained within a few passages. In some cases, tumor-derived organoids were re-implanted into the dorsal skin.
Western blotting
Organoids were harvested after Cell Recovery Solution (BD Biosciences) treatment for 1 h on ice and lysed using RIPA buffer (50 mM Tris-HCl at pH 7.4 supplemented with 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and 1 mM EDTA) supplemented with 1 M NaF, 0.1 M Na3VO4, 1 M βGP, and a protease inhibitor cocktail (Nacalai Tesque, Kyoto, Japan). Protein concentrations were quantified using a Pierce BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA), and 5–10 µg of protein was separated under reducing conditions in regular tris-glycine buffer on a gradient gel, SuperSep Ace 5 20% (Wako). The proteins were transferred onto Immobilon-P PVDF membranes (Sigma-Aldrich) using semi-dry electrophoresis. The membrane was blocked with PBS-T containing 5% non-fat dry milk at 4 °C overnight, followed by incubation with primary antibodies for 90 min at room temperature. The antibodies used were as follows: RasG12D (#14429, Cell Signaling Technology, Danverse, MA, 1:1,000), p16Ink4a (sc-1207; Santa Cruz Biotechnology, Dallas, TX, 1:2,000), Pten (#9559, Cell Signaling Technology, 1:2,000), p53 (#2524, Cell Signaling Technology, 1:2,000), p19Arf (NB200-174, Novus Biologicals, Centennial, CO, 1:1,000), Akt (#4961, Cell Signaling Technology, 1:2,000), pAkt (#4060, Cell Signaling Technology, 1:2,000), α-tubulin (T5168, Sigma-Aldrich, 1:20,000), and β-actin (013–24553, Wako, 1:2,000). The membranes were incubated with the corresponding secondary antibodies (mouse: NA931, GE Healthcare, Buchinghamshire, UK, rabbit: 7074, Cell Signaling Technology) for 45 min at room temperature and visualized using ImmunoStar LD (Wako). Chemiluminescent images were captured using an ImageQuant LAS 4000 mini (GE Healthcare).
Histopathological analysis
All tumors were fixed in 10–20% buffered neutral formalin, dehydrated, and embedded in paraffin. For the organoids, Matrigel was lysed by treatment with Cell Recovery Solution (BD Biosciences) and embedded in iPGell (GenoStaff, Tokyo, Japan) before fixation. The formalin-fixed, paraffin-embedded samples were sliced into 3-µm-thick sections and subjected to hematoxylin and eosin (H&E) and immunohistochemical staining with the following antibodies: pan cytokeratin clone AE1/AE3 (ab27988, Abcam, Tokyo, Japan, 1:40) and vimentin clone EPR3776 (#2707-1, Epitomics, Burlingame, CA, 1:500). The reactions were visualized using the Dako REAL EnVision Detection System (Dako, Glostrup, Denmark) with diaminobenzidine chromogen as the substrate.
Array-based comparative genomic hybridization analysis
To extract genomic DNA from organoids, a NucleoSpin Tissue Kit (Takara, Shiga, Japan) was used. DNA quantity and quality was assessed using a NanoDrop spectrophotometer and agarose gel electrophoresis (Thermo Fisher Scientific), respectively. A SureTaq DNA Labeling Kit (5190-03399, Agilent, Santa Clara, CA) was used to chemically label 500 ng of genomic DNA with either ULS-Cy5 or ULS-Cy3 dye. Hybridization with labeled DNA was performed using a SurePrint G3 mouse CGH microarray 4 × 180 K (G4826A, Agilent). Scanning and image analysis were performed using Agilent Feature Extraction ver.11.0 (Agilent) and a SureScan Microarray Scanner (G4900DA, Agilent). Agilent Genomic Workbench ver.7.0.4.0 software was used to visualize, detect, and analyze chromosomal patterns in the microarray profiles. Organoids without lentiviral infection were analyzed as reference for estimating copy number variations.
Transcriptome analysis
Total RNA was extracted using an RNeasy Mini Kit (Qiagen). The RNA quality and quantity of each sample were checked and measured, respectively, using an Agilent 2100 Bioanalyzer (Agilent). All samples had an RNA integrity number greater than 7.8. The cRNA was prepared from 200 ng of total RNA using a Low Input Quick Amp Labeling Kit, one-color (5190-2305, Agilent), and labeled with cyanin3. Hybridization was performed using the SurePrint G3 Mouse Gene Exp v2 Array Kit 8 × 60 K (G4852B, Agilent). Scanning and image analysis were performed on a SureScan Microarray Scanner (G4900DA, Agilent). Microarray data were analyzed using GeneSpring GX ver.13.1 software (Agilent). Organoids without lentiviral infection served as reference. Data are available at the GEO database (GSE175512).
Supplementary information
Acknowledgements
This work was supported in part by the Japan Society for the Promotion of Science (grant number 17K19624, 21K09483, and 21K09506) and a research grant from Chiba Prefecture. We thank N. Sakurai, K. Takahashi, K. Fujiwara, N. Miyazawa, A. Washio, and M. Kohno for their excellent technical assistance. We are also grateful to the animal experiment facility at Chiba Cancer Center. We thank Elaine Fuchs for providing LV-Cre pLKO.1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41389-021-00337-8.
References
- 1.Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170:17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–67. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arner EN, Du W, Brekken RA. Behind the wheel of epithelial plasticity in KRAS-driven cancers. Front Oncol. 2019;9:1049. doi: 10.3389/fonc.2019.01049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drosten M, Guerra C, Barbacid M. Genetically engineered mouse models of K-Ras-driven lung and pancreatic tumors: validation of therapeutic targets. Cold Spring Harb Perspect Med. 2018;8:8. doi: 10.1101/cshperspect.a031542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 6.Yamagami W, Nagase S, Takahashi F. Clinical statistics of gynecologic cancers in Japan. J Gynecol Oncol. 2017;28:e32. doi: 10.3802/jgo.2017.28.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research N, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murali R, Soslow RA, Weigelt B. Classification of endometrial carcinoma: more than two types. Lancet Oncol. 2014;15:e268–278. doi: 10.1016/S1470-2045(13)70591-6. [DOI] [PubMed] [Google Scholar]
- 9.Podsypanina K, Ellenson LH, Nemes A, Gu J, Tamura M, Yamada KM, et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci USA. 1999;96:1563–8. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stambolic V, Tsao MS, Macpherson D, Suzuki A, Chapman WB, Mak TW. High incidence of breast and endometrial neoplasia resembling human Cowden syndrome in pten+/- mice. Cancer Res. 2000;60:3605–11. [PubMed] [Google Scholar]
- 11.Daikoku T, Hirota Y, Tranguch S, Joshi AR, DeMayo FJ, Lydon JP, et al. Conditional loss of uterine Pten unfailingly and rapidly induces endometrial cancer in mice. Cancer Res. 2008;68:5619–27. doi: 10.1158/0008-5472.CAN-08-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng H, Liu P, Zhang F, Xu E, Symonds L, Ohlson CE, et al. A genetic mouse model of invasive endometrial cancer driven by concurrent loss of Pten and Lkb1 Is highly responsive to mTOR inhibition. Cancer Res. 2014;74:15–23. doi: 10.1158/0008-5472.CAN-13-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joshi A, Ellenson LH. Adenovirus mediated homozygous endometrial epithelial Pten deletion results in aggressive endometrial carcinoma. Exp Cell Res. 2011;317:1580–9. doi: 10.1016/j.yexcr.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim TH, Wang J, Lee KY, Franco HL, Broaddus RR, Lydon JP, et al. The synergistic effect of conditional Pten loss and oncogenic K-ras mutation on endometrial cancer development occurs via decreased progesterone receptor action. J Oncol. 2010;2010:139087. doi: 10.1155/2010/139087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tirodkar TS, Budiu RA, Elishaev E, Zhang L, Mony JT, Brozick J, et al. MUC1 positive, Kras and Pten driven mouse gynecologic tumors replicate human tumors and vary in survival and nuclear grade based on anatomical location. PLoS ONE. 2014;9:e102409. doi: 10.1371/journal.pone.0102409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wild PJ, Ikenberg K, Fuchs TJ, Rechsteiner M, Georgiev S, Fankhauser N, et al. p53 suppresses type II endometrial carcinomas in mice and governs endometrial tumour aggressiveness in humans. EMBO Mol Med. 2012;4:808–24. doi: 10.1002/emmm.201101063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 18.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–72. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 19.Bartfeld S, Bayram T, van de Wetering M, Huch M, Begthel H, Kujala P, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148:126–.e126. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen YW, Huang SX, de Carvalho A, Ho SH, Islam MN, Volpi S, et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat Cell Biol. 2017;19:542–9. doi: 10.1038/ncb3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schumacher MA, Aihara E, Feng R, Engevik A, Shroyer NF, Ottemann KM, et al. The use of murine-derived fundic organoids in studies of gastric physiology. J Physiol. 2015;593:1809–27. doi: 10.1113/jphysiol.2014.283028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onuma K, Ochiai M, Orihashi K, Takahashi M, Imai T, Nakagama H, et al. Genetic reconstitution of tumorigenesis in primary intestinal cells. Proc Natl Acad Sci USA. 2013;110:11127–32. doi: 10.1073/pnas.1221926110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato T, Morita M, Tanaka R, Inoue Y, Nomura M, Sakamoto Y, et al. Ex vivo model of non-small cell lung cancer using mouse lung epithelial cells. Oncol Lett. 2017;14:6863–8. doi: 10.3892/ol.2017.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato S, Fushimi K, Yabuki Y, Maru Y, Hasegawa S, Matsuura T, et al. Precision modeling of gall bladder cancer patients in mice based on orthotopic implantation of organoid-derived tumor buds. Oncogenesis. 2021;10:33. doi: 10.1038/s41389-021-00322-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochiai M, Yoshihara Y, Maru Y, Matsuura T, Izumiya M, Imai T, et al. Kras-driven heterotopic tumor development from hepatobiliary organoids. Carcinogenesis. 2019;40:1142–52. doi: 10.1093/carcin/bgz024. [DOI] [PubMed] [Google Scholar]
- 26.Matsuura T, Maru Y, Izumiya M, Hoshi D, Kato S, Ochiai M, et al. Organoid-based ex vivo reconstitution of Kras-driven pancreatic ductal carcinogenesis. Carcinogenesis. 2020;41:490–501. doi: 10.1093/carcin/bgz122. [DOI] [PubMed] [Google Scholar]
- 27.Maru Y, Tanaka N, Ebisawa K, Odaka A, Sugiyama T, Itami M, et al. Establishment and characterization of patient-derived organoids from a young patient with cervical clear cell carcinoma. Cancer Sci. 2019;110:2992–3005. doi: 10.1111/cas.14119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–8. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cherniack AD, Shen H, Walter V, Stewart C, Murray BA, Bowlby R, et al. Integrated molecular characterization of uterine carcinosarcoma. Cancer Cell. 2017;31:411–23. doi: 10.1016/j.ccell.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McConechy MK, Ding J, Cheang MC, Wiegand K, Senz J, Tone A, et al. Use of mutation profiles to refine the classification of endometrial carcinomas. J Pathol. 2012;228:20–30. doi: 10.1002/path.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 32.Liang X, Daikoku T, Terakawa J, Ogawa Y, Joshi AR, Ellenson LH, et al. The uterine epithelial loss of Pten is inefficient to induce endometrial cancer with intact stromal Pten. PLoS Genet. 2018;14:e1007630. doi: 10.1371/journal.pgen.1007630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He XC, Yin T, Grindley JC, Tian Q, Sato T, Tao WA, et al. PTEN-deficient intestinal stem cells initiate intestinal polyposis. Nat Genet. 2007;39:189–98. doi: 10.1038/ng1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marsh V, Winton DJ, Williams GT, Dubois N, Trumpp A, Sansom OJ, et al. Epithelial Pten is dispensable for intestinal homeostasis but suppresses adenoma development and progression after Apc mutation. Nat Genet. 2008;40:1436–44. doi: 10.1038/ng.256. [DOI] [PubMed] [Google Scholar]
- 35.Memarzadeh S, Zong Y, Janzen DM, Goldstein AS, Cheng D, Kurita T, et al. Cell-autonomous activation of the PI3-kinase pathway initiates endometrial cancer from adult uterine epithelium. Proc Natl Acad Sci USA. 2010;107:17298–303. doi: 10.1073/pnas.1012548107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leskela S, Pérez-Mies B, Rosa-Rosa JM, Cristobal E, Biscuola M, Palacios-Berraquero ML, et al. (2019). Molecular Basis of Tumor Heterogeneity in Endometrial Carcinosarcoma. Cancers (Basel)11. [DOI] [PMC free article] [PubMed]
- 37.Tang FH, Hsieh TH, Hsu CY, Lin HY, Long CY, Cheng KH, et al. KRAS mutation coupled with p53 loss is sufficient to induce ovarian carcinosarcomas in mice. Int J Cancer. 2017;140:1860–9. doi: 10.1002/ijc.30591. [DOI] [PubMed] [Google Scholar]
- 38.Bailey JM, Hendley AM, Lafaro KJ, Pruski MA, Jones NC, Alsina J, et al. p53 mutations cooperate with oncogenic Kras to promote adenocarcinoma from pancreatic ductal cells. Oncogene. 2016;35:4282–8. doi: 10.1038/onc.2015.441. [DOI] [PubMed] [Google Scholar]
- 39.O’dell MR, Huang JL, Whitney-Miller CL, Deshpande V, Rothberg P, Grose V, et al. Kras(G12D) and p53 mutation cause primary intrahepatic cholangiocarcinoma. Cancer Res. 2012;72:1557–67. doi: 10.1158/0008-5472.CAN-11-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCluggage WG. Malignant biphasic uterine tumours: carcinosarcomas or metaplastic carcinomas? J Clin Pathol. 2002;55:321–5. doi: 10.1136/jcp.55.5.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao S, Bellone S, Lopez S, Thakral D, Schwab C, English DP, et al. Mutational landscape of uterine and ovarian carcinosarcomas implicates histone genes in epithelial-mesenchymal transition. Proc Natl Acad Sci USA. 2016;113:12238–43. doi: 10.1073/pnas.1614120113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cuevas IC, Sahoo SS, Kumar A, Zhang H, Westcott J, Aguilar M, et al. Fbxw7 is a driver of uterine carcinosarcoma by promoting epithelial-mesenchymal transition. Proc Natl Acad Sci USA. 2019;116:25880–90. doi: 10.1073/pnas.1911310116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gotoh O, Sugiyama Y, Takazawa Y, Kato K, Tanaka N, Omatsu K, et al. Clinically relevant molecular subtypes and genomic alteration-independent differentiation in gynecologic carcinosarcoma. Nat Commun. 2019;10:4965. doi: 10.1038/s41467-019-12985-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burgess MR, Hwang E, Mroue R, Bielski CM, Wandler AM, Huang BJ, et al. KRAS allelic imbalance enhances fitness and modulates MAP kinase dependence in cancer. Cell. 2017;168:817–.e815. doi: 10.1016/j.cell.2017.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z, Wang Y, Vikis HG, Johnson L, Liu G, Li J, et al. Wildtype Kras2 can inhibit lung carcinogenesis in mice. Nat Genet. 2001;29:25–33. doi: 10.1038/ng721. [DOI] [PubMed] [Google Scholar]
- 46.Boretto M, Maenhoudt N, Luo X, Hennes A, Boeckx B, Bui B, et al. Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat Cell Biol. 2019;21:1041–51. doi: 10.1038/s41556-019-0360-z. [DOI] [PubMed] [Google Scholar]
- 47.Maru Y, Hippo Y. Current status of patient-derived ovarian cancer models. Cells. 2019;8:8. doi: 10.3390/cells8050505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maru Y, Tanaka N, Itami M, Hippo Y. Efficient use of patient-derived organoids as a preclinical model for gynecologic tumors. Gynecol Oncol. 2019;154:189–98. doi: 10.1016/j.ygyno.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 49.Maru Y, Kawata A, Taguchi A, Ishii Y, Baba S, Mori M, et al. (2020). Establishment and molecular phenotyping of organoids from the squamocolumnar junction region of the uterine cervix. Cancers (Basel). (2020);12:694. [DOI] [PMC free article] [PubMed]
- 50.Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- 51.Maru Y, Orihashi K, Hippo Y. Lentivirus-based stable gene delivery into intestinal organoids. Methods Mol Biol. 2016;1422:13–21. doi: 10.1007/978-1-4939-3603-8_2. [DOI] [PubMed] [Google Scholar]
- 52.Beronja S, Livshits G, Williams S, Fuchs E. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nat Med. 2010;16:821–7. doi: 10.1038/nm.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.