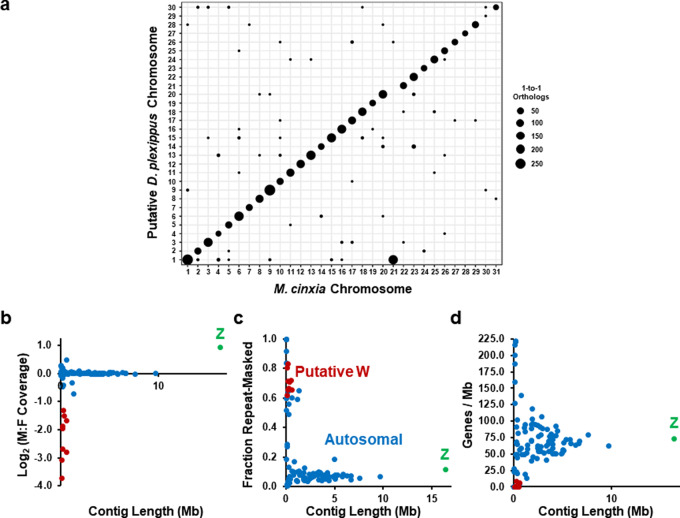
Fig. 1. Assignation of the DpMex_v1 genome assembly to the chromosomes of D. plexippus.
a Chromosomal anchoring of contigs based on the chromosome of M. cinxia showing the highest number of 1-to-1 orthologs for protein-coding genes in our OGS1_DpMex. Overall, 97.02% (4,855 out of 5,004) orthologs mapped to chromosomes of M. cinxia provided coherent cytological information about to which chromosomes the contigs should be anchored. The diameter of the circles denotes the number of such 1-to-1 orthologs. Overall, high synteny conservation between the two species can be observed, which supports that the gene content of the ancestral chromosomal elements to these two species remained essentially intact. It can also be observed that chromosome 1 in D. plexippus encompasses genes from chromosomes 1 and 21 of M. cinxia, reflecting the outcome of a fusion event. For each of the 108 contigs part of the DpMex_v1 assembly, (b) the log2 of the male to female coverage, (c) the repeat-masked fraction, and (d) gene density, is plotted against its length. A color code is used to distinguish the contigs categorized as autosomal, Z-related, and potentially W-related. The raw data for these plots are provided in Supplementary Data 3.