Abstract
Background
Effective tiller number (ETN) has a pivotal role in determination of rice (Oryza sativa L.) grain yield. ETN is a complex quantitative trait regulated by both genetic and environmental factors. Despite multiple tillering-related genes have been cloned previously, few of them have been utilized in practical breeding programs.
Results
In this study, we conducted a genome-wide association study (GWAS) for ETN using a panel of 490 rice accessions derived from the 3 K rice genomes project. Thirty eight ETN-associated QTLs were identified, interestingly, four of which colocalized with the OsAAP1, DWL2, NAL1, and OsWRKY74 gene previously reported to be involved in rice tillering regulation. Haplotype (Hap) analysis revealed that Hap5 of OsAAP1, Hap3 and 6 of DWL2, Hap2 of NAL1, and Hap3 and 4 of OsWRKY74 are favorable alleles for ETN. Pyramiding favorable alleles of all these four genes had more enhancement in ETN than accessions harboring the favorable allele of only one gene. Moreover, we identified 25 novel candidate genes which might also affect ETN, and the positive association between expression levels of the OsPILS6b gene and ETN was validated by RT-qPCR. Furthermore, transcriptome analysis on data released on public database revealed that most ETN-associated genes showed a relatively high expression from 21 days after transplanting (DAT) to 49 DAT and decreased since then. This unique expression pattern of ETN-associated genes may contribute to the transition from vegetative to reproductive growth of tillers.
Conclusions
Our results revealed that GWAS is a feasible way to mine ETN-associated genes. The candidate genes and favorable alleles identified in this study have the potential application value in rice molecular breeding for high ETN and grain yield.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12284-021-00495-8.
Keywords: Effective tiller number, GWAS, Rice
Background
Rice (Oryza sativa L.) is one of the main staple crops worldwide and improving rice yield is an urgent need for the increasing world’s population (Lobell et al. 2011). Effective tiller number (ETN) per plant is an essential yield component for rice and affected by both genetic and environmental factors (Xing & Zhang 2010). Rice tiller derives from two processes, axillary meristem (AM) formation and tiller bud outgrowth (Wang & Li 2011), which are regulated by many genes. MOC1, encoding a GRAS domain transcription factor, plays a pivotal role in AM formation, and the loss-of-function moc1 mutant shows a monoculm phenotype (Li et al. 2003). MOC3/OsWUS/TILLERS ABSENT1 (TAB1) is also indispensable for tiller bud formation (Lu et al. 2015b), which may promote AM initiation by inducing the expression of OSH1 (Tanaka et al. 2015). Besides, LAX PANICLE1 (LAX1) and LAX2 can physically interact with MOC1 and also regulate AM formation (Oikawa & Kyozuka 2009; Tabuchi et al. 2011). Tiller bud outgrowth is suppressed by a newly discovered plant hormone, strigolactones (SLs) (Gomez-Roldan et al. 2008). Typical SL mutants display dwarf and increased tillering, such as d27 (Lin et al. 2009), htd1/d17 (Zou et al. 2006), and d10 (Arite et al. 2007; Yuan et al. 2013), whose responsible genes are involved in SL synthesis; htd2/d14 (Liu et al. 2009), d3 (Yasuno et al. 2009; Zhao et al. 2014) and d53 (Zhou et al. 2013; Jiang et al. 2013), whose responsible genes are involved in SL signaling.
Besides SL, other plant hormones can also affect rice tiller growth. For instance, overexpression of OsPIN2 or OsPIN9, both encoding an auxin efflux transporter, led to increased tiller number (TN) (Chen et al. 2012; Hou et al. 2021). OsCKX9, encoding a cytokinin (CK) catabolic enzyme, suppressed tillering upon transcriptional activation by SL (Duan et al. 2019). DELLA protein SLENDER RICE 1, a repressor of gibberellin (GA) signaling, inhibits MOC1 degradation to regulate both tillering and plant height (Liao et al. 2019). Tiller growth could also be influenced by some genes independent of plant hormones. For example, MOC2 encodes a fructose-1,6-bisphosphatase participating in sucrose synthesis, and the moc2 mutant also shows monoculm phenotype (Koumoto et al. 2013). Hd3a is the homolog of Arabidopsis FLOWERING LOCUST (FT) protein in rice, which is transported from the phloem to shoot apical cells and promotes lateral branching (Zhao et al. 2015). The rcn1 mutant displays monoculm phenotype and the responsible gene OsABCG5 encodes an ATP-binding cassette protein required for rice shoot branching (Yasuno et al. 2009).
Fast-growing next-generation sequencing (NGS) technology has become a cheaper solution for genotyping, which makes it possible to use high-throughput single nucleotide polymorphism (SNP) markers to perform GWAS (Liu & Yan 2019). Many genes controlling important agronomic traits have been identified using this method in recent years, such as OsSPL13 controlling rice grain size (Si et al. 2016), OsNPF6.1 associated with nitrogen use efficiency in rice (Tang et al. 2019), ZmVPP1 contributing to drought tolerance in maize (Wang et al. 2016), and ZmFBL41 conferring banded leaf and sheath blight resistance in maize (Li et al. 2019). As an essential yield-related trait, rice TN has also been studied by GWAS. A previous study on 14 agronomic traits had identified eight loci associated with TN, distributing on chromosome 1, 2, 4 and 10 (Huang et al. 2010). In another study on 15 agronomic traits using a high-throughput phenotyping facility, an F-box gene OsFBL20 controlling TN was identified (Yang et al. 2014). A recent GWAS identified 15 novel loci associated with TN variations, and five candidate genes were validated (Jiang et al. 2019). Another study based on TN data in different stages revealed that dynamic change in TN played a key role in determination of panicle number and identified a new gene OsSAUR27 associated with TN (Ma et al. 2020).
Most genes regulating rice tillering have been cloned from high- or low-tillering mutants which are rarely used as rice breeding materials. However, ETN is a complex trait with a relatively low heritability and contributed by multiple QTLs (Liu et al. 2010). Therefore, in order to clone rice tillering genes potentially used by breeders, more QTLs need to be identified from the natural rice population. In this study, we performed GWAS based on ETN data of a panel of 490 rice accessions grown in two locations, and the objectives are as follows: (1) feasibility analysis of mining ETN genes by GWAS; (2) identify novel genes affecting ETN; (3) probe favorable alleles of ETN-associated genes.
Methods
Plant Materials
The 490 rice accessions used in this study were derived from the 3000 Rice Genomes Project (Wang et al. 2018). Information of these rice accessions was shown in Additional file 1: Table S1.
Phenotyping Analysis
For the field experiments, rice seeds were submerged in clean water at 37 °C for 48 h and transferred to nursery bed for germination. One-month-old seedlings were then transplanted into the paddy field at density of 16 cm × 20 cm and one plant per hill. A completely randomized block design with three replicates was performed. Six plants in the middle of every row were selected for evaluation of ETN with effective tiller being defined as the one bearing more than 10 seeds at maturity stage. The mean value of three replicates was used for analysis. The field experiment was performed in two different years and locations: (1) June to November 2017, Fengcheng city, Jiangxi (JX) Province, China (28°15′N, 115°77′E); (2) December 2017 to April 2018, Sanya city, Hainan (HN) Province, China (18°25′N, 109°51′E). Broad-sense heritability (H2) was calculated based on the following formular: H 2 = VG/(VG + VE) , where VG and VE are genetic and environmental variances.
Genotyping Data and SNP Filtering
The raw genotype data of the 490 accessions were obtained from the Rice SNP-Seek Database (https://snp-seek.irri.org/) (Alexandrov et al. 2015). A total of 5,877,569, 4,542,091 and 4,995,512 cleaned SNPs were called in the whole, indica and non-indica (nonind) population, respectively. SNPs were filtered utilizing the software PLINK (Purcell et al. 2007) with missing rate < 40% and minor allele frequency (MAF) > 0.05.
Population Structure and Phylogenetic Analysis
The software ADMIXTURE 1.3 (Alexander et al. 2009) was used to calculate the population structure. A total of 394,572 SNPs pruned by PLINK with r2 > 0.3 were used for the analysis. The ancestral population number (K) was assumed ranging from 2 to 6. Principal component analysis (PCA) was performed using PLINK and plotted with R ‘ggplot2’ package (Wickham. 2016). The FastTree software (Price et al. 2009) was used to construct the phylogenetic tree based on the approximately maximum-likelihood method, and the generated Newick file was then visualized on the iTOL website (https://itol.embl.de/) (Letunic & Bork. 2019).
GWAS and Linkage Disequilibrium (LD) Analysis
GWAS was performed with EMMAX program (Kang et al. 2010). Briefly, we first used PLINK to get the genotype file, and the ETN phenotype data were collected from two locations, Jiangxi and Hainan. The genotype and phenotype file, as well as the Balding-Nichols kinship matrix were used to fit the mixed linear model in EMMAX. The Manhattan and quantile-quantile (Q-Q) plots were generated using the R package “qqman” (Turner. 2018). The pairwise linkage disequilibrium (LD) (r2) was calculated using PopLDdecay software (Zhang et al. 2019). The Pairwise r2 was calculated for the SNPs in 500 kb and averaged in 1 kb across the whole genome. The LD decay rate was measured as the chromosomal distance at which the average pairwise correlation coefficient (r2) decreased to half of its maximum value (Huang et al. 2010). The regional Manhattan plot and LD heatmap were obtained using LDBlockShow software (Dong et al. 2020).
Haplotype Analysis
The haplotypes of candidate genes were classified using SNPs and InDels (insertion and deletion) in the promoter (2 kb upstream of ATG) and untranslated region (UTR), as well as non-synonymous SNPs in the coding sequence (CDS). Haplotypes with more than 10 accessions were retained to perform one-way variance analysis (ANOVA) in R. Multiple comparisons were conducted using R package “agricolae”.
Resequencing of OsPILS6b
Genomic DNA of 28 accessions (Additional file 1: Table S1) was extracted using standard CTAB method (Doyle 1987), and the 5.9 kb full-length sequence for the OsPILS6b gene (including 2.5 kb upstream of ATG) was amplified with high-fidelity DNA polymerase KOD FX (TOYOBO, KFX-101) using the following primers, PILS6bF: CAGTCAAGCATCTCACCCTTT, PILS6bR: AGCCGATTGGTTTAT ACTGGA. PCR product was sequenced directly by the Beijing Genomics Institute. Multiple sequence alignment was performed using MEGA X (Sudhir et al. 2018).
Reverse Transcription Quantitative-PCR Analysis
Eight, twelve, and eight accessions representing three different haplotypes of OsPILS6b were selected from the 490 rice panel (Additional file 1: Table S1), respectively. Shoot bases (about 0.5 cm) of 10-day-old rice seedlings were used for total RNA isolation using TRIzol reagent (Invitrogen). One μg of RNA was treated with DNase I (TAKARA) and then used for cDNA synthesis (TAKARA) according to the manufacturer’s instructions. The rice OsActin1 gene (LOC_Os03g50885) was used as an internal control. The reverse transcription quantitative-PCR (RT-qPCR) primers used for OsActin1 and OsPILS6b were as follows: F1: TCCATCTTGGCATCTCTCAG, R1: GGTACCCTCATCAGGCATCT; F2: ACCTTTGACAGCTGCGATGA, R2: ATAGCAGGGGCTCTTCCTCA. The qRT-PCR was performed on an ABI Prism 7500 instrument (Applied Biosystems) and relative gene expression level was calculated using the 2-ΔΔCT method (Livak & Schmittgen 2001).
Results
Population Structure and ETN Variation of the 490 Rice Accessions
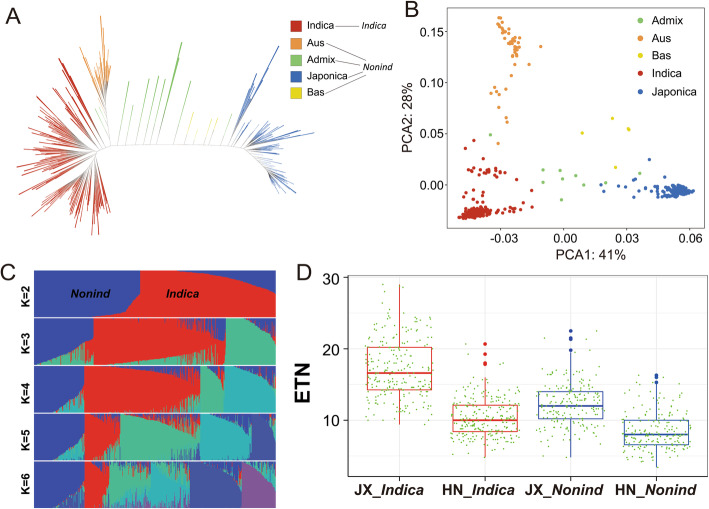
The 490 rice accessions consist of 235 Indica, 194 Japonica, 45 Aus, 11 Admixture (Admix) and 5 Basmati (Bas), according to the K9_group_Admixture (Wang et al. 2018) (Additional file 1: Table S1). Due to the smaller population size compared with that of Indica, for the follow-up analysis, Japonica, Aus, Admix and Bas were combined into one group named as non-indica (nonind). Phylogenic tree (Fig. 1a) and population structure analysis (Fig. 1c, Additional file 5: Fig. S1a) demonstrate that these two subpopulations were distinguishable according to their genotype, which was also supported by result of PCA analysis (Fig. 1b). Linkage disequilibrium (LD) analysis showed the LD decay rate was higher in indica than nonind (Additional file 5: Fig. S1b), consistent with previous studies (Huang et al. 2010; Wang et al. 2018).
Fig. 1.
Population structure and ETN variation of 490 rice accessions. (a) Phylogenetic tree constructed using LD-pruned SNPs. (b) PCA plots for the first two components of 490 rice accessions. (c) Population structure determined by ADMIXTURE. Ancestral population number (K) ranged from two to six. (d) ETN variation of two subpopulations in two locations. Boxplot described ETN distribution of one subpopulation in one location, and each dot around the boxplot represent ETN value of one accession. ETN, effective tiller number; JX, Jiangxi province; HN, Hainan province. Different letters denote significant differences (P < 0.05) based on Duncan’s multiple-range test
This panel of 490 rice accessions was grown in summer 2017 in Jiangxi Province (JX) and winter 2017 in Hainan Province (HN), respectively. ETN variation was quite abundant in our population, ranging from 4.8 to 30 and 3.4 to 20.7 in JX and HN, respectively. Overall, indica accessions showed more ETN and wider ETN range than nonind accessions, similar to the trend observed in a previous study (Ma et al. 2020). The results also showed that both indica and nonind accessions had more tillers in JX than in HN (Fig. 1d). We speculated that the short daylight condition in winter in HN promotes flowering and restrains the vegetative growth and tillering in rice. Despite the dramatic environmental difference, ETN in these two locations were slightly correlated, with a correlation coefficient of R2 = 0.45. Broad-sense heritability (H2) of ETN was 0.58, consistent with a previous study (Liu et al. 2010). The relatively low heritability indicates that besides genetic factors, environmental factors also play a critical role in ETN variation.
Overview of QTLs Associated with ETN Detected by GWAS
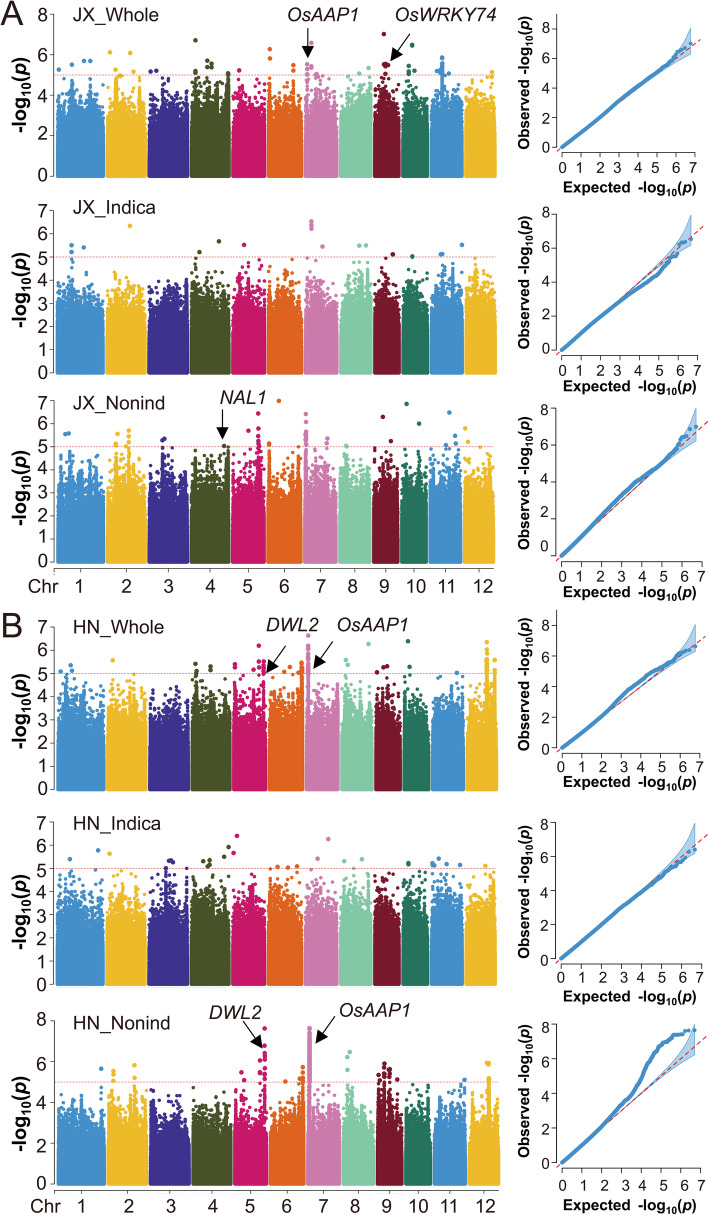
As mentioned above, both the population and the location have significant effect on ETN. Therefore, to exclude effect of population structure and environments, we performed six GWAS assays depending on the location and subpopulation, i.e., the whole, indica and nonind populations in JX and HN, respectively. As reported previously, a QTL was called when there were at least two significant SNPs (P ≤ 10E-5) within 200-kb range (Jiang et al. 2019), and the region of a specific QTL was defined as 300 kb flanking the lead SNP (SNP with the lowest P value in a cluster) (Guo et al. 2020). By this standard, a total of 38 ETN-associated QTLs were identified (Fig. 2), among which 7 QTLs were detected by two or three GWAS assays. Detailed information of these QTLs was listed in Additional file 2: Table S2. A total of 1134 genes are located in these regions.
Fig. 2.
Manhattan plots and quantile-quantile (Q-Q) plots of six GWAS assays. (a) GWAS assays for the whole, indica and nonind population using ETN data in Jiangxi, (b) GWAS assays for whole, indica and nonind population using ETN data in Hainan. Some candidate genes are indicated using arrow heads
Colocalization of ETN-Associated QTLs with Previously Reported Rice Tillering Genes
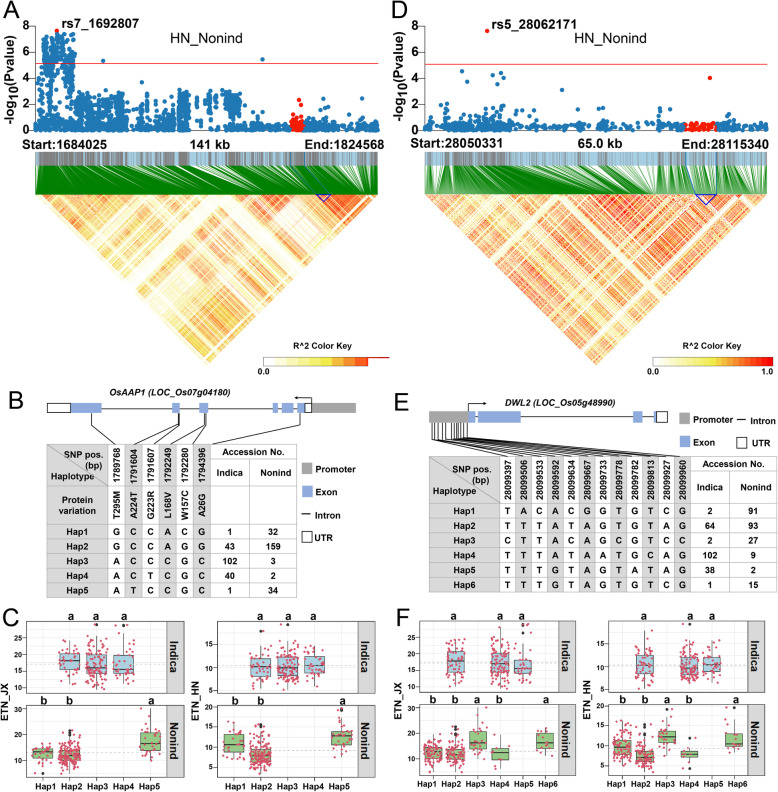
To evaluate the reliability of our GWAS results, we examined whether the detected ETN-associated QTLs could colocalize with some known genes involved in rice tillering. First, we targeted ETN7–3, which was detected in three GWAS assays (JX_whole, HN_whole, and HN_nonind) and showed the most remarkable signal (Fig. 2). Fortunately, the OsAAP1 gene encoding an amino acid transporter which has recently been reported to affect TN, is located in this region. Overexpression lines of OsAAP1 showed enhanced TN, whereas RNAi lines showed opposite phenotypes (Ji et al. 2020). OsAAP1 was located around 102 kb downstream of the peak SNP (rs7_1692807) of ETN7–3. Therefore, we called all the SNPs within 141-kb region flanking OsAAP1 and investigated the linkage disequilibrium (LD) of OsAAP1 in the 490 rice accessions panel. The LD heatmap showed that OsAAP1 is located in a region with relatively high LD (Fig. 3a). Next, we analyzed the haplotype of OsAAP1 in both indica and nonind subpopulations. In total, we identified six non-synonymous SNPs in the coding sequence (CDS) (Fig. 3b). Based on these variants, we identified 5 haplotypes of OsAAP1. The indica and nonind subgroup have three different major haplotypes, respectively, and Hap2 is present as a major haplotype in both subgroup. In agreement with the fact that OsAAP1 was detected in nonind rather than indica subgroup, there seems no much difference in ETN among the three major haplotypes (Hap2, 3, 4) in indica subgroup. However, the ETN varies significantly among the three major haplotypes (Hap1, 2 and 5) in nonind subgroup, with Hap5 showing the highest ETN in both JX and HN (Fig. 3c). Therefore, Hap5 is a favorable allele for ETN and introducing this Hap5 allele of OsAAP1 into other cultivars may contribute to an increase of ETN.
Fig. 3.
Associated region and haplotype analysis of OsAAP1 and DWL2. (a, d) Regional Manhattan plots and LD heatmap of OsAAP1 (a) and DWL2 (d). The single red dot above the red line denotes the lead SNP, the red dots under the red line and the blue triangle in the LD heatmap denote the genomic region of candidate genes. (b, e) Gene structure and marker information of OsAAP1 (b) and DWL2 (e). Arrow heads indicate the direction of gene transcription. (c, f) Haplotype analysis of OsAAP1 (c) and DWL2 (f). Boxplot described ETN distribution of one subpopulation with one haplotype, and each red dot around the boxplot represents ETN value of one accession. The gray dotted-lines denote average ETN of one subgroup. Different letters denote significant differences (P < 0.05) based on Duncan’s multiple-range test
Another noticeable signal was ETN5–5, which was also detected by both nonind subpopulation and the whole population (HN_nonind and HN_whole) (Fig. 2b). A WUSCHEL-related homeobox (WOX) transcription factor gene DWT1-LIKE2 (DWL2) was thought to be the candidate. The previous report has shown that DWARF TILLER 1 (DWT1), a WOX transcription factor, act as a positive regulator of tiller growth (Wang et al. 2014) and a DWT1 homolog DWL2 shares partial functional redundancy with DWT1 in controlling uniform growth of rice tillers and main shoot (Fang et al. 2020). DWL2 was located 38 kb downstream of the lead SNP (rs5_28062171). LD heatmap showed a moderate LD level around the DWL2 gene (Fig. 3d). A total of 12 SNPs were detected in the promoter region, while no non-synonymous SNP or InDel was found in CDS (Fig. 3e). Haplotype analysis revealed that Hap3 and Hap6 are favorable alleles for ETN and preferentially exist in nonind subgroup (Fig. 3f). Like OsAAP1, these two alleles of DWL2 from nonind subgroup may also be useful for enhancement of ETN when introduced to Indica cultivars.
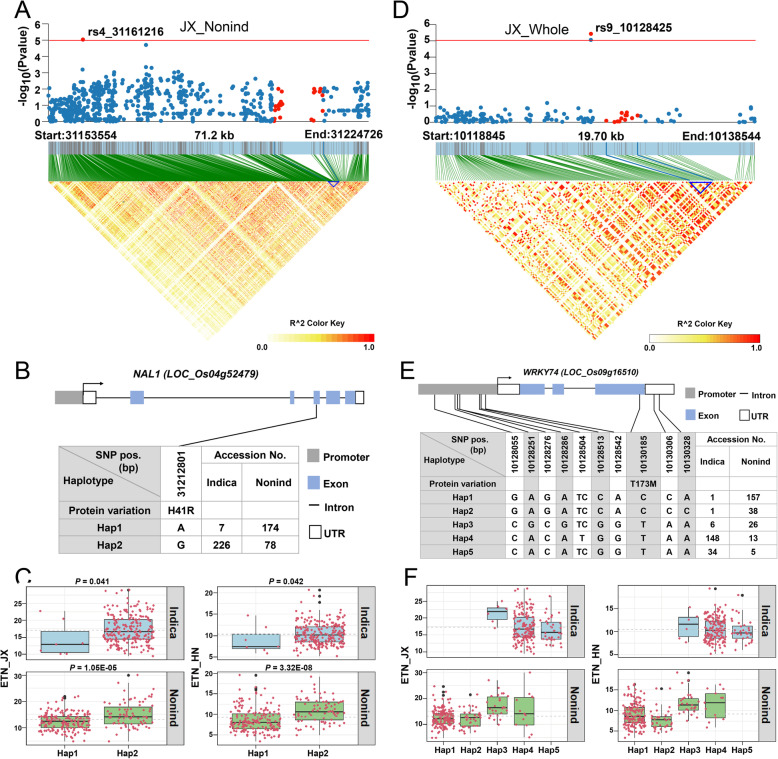
Within the QTL ETN4–5 detected in JX_nonind population (Fig. 2a), we identified the Narrow Leaf1 (NAL1) gene, which was initially cloned from a loss-of-function mutant showing narrow leaf and increased TN (Qi et al. 2008; Jiang et al. 2015). We identified a non-synonymous SNP that was previously reported to be associated with panicle number (Yano et al. 2016) (Fig. 4b). The results demonstrated that Hap2 with G allele showed more ETN than Hap1 with A allele in both indica and nonind subgroup (Fig. 4c). Furthermore, this SNP was also identified in a major QTL LSCHL4 from Japonica cultivar, which can increase grain yield when introduced into the Indica super rice variety 93–11 (Zhang et al. 2014).
Fig. 4.
Associated region and haplotype analysis of NAL1 and OsWRKY74. (a, d) Regional Manhattan plots and LD heatmap of NAL1 (a) and OsWRKY7 (c). The single red dot above the red line denotes the lead SNP, the red dots under the red line and the blue triangle in the LD heatmap denote the genomic region of candidate genes. (b, e) Gene structure and marker information of NAL1 (b) and OsWRKY74 (e). Arrow heads indicate the direction of gene transcription. (c, f) Haplotype analysis of OsAAP1 (c) and OsWRKY74 (f). Boxplot described ETN distribution of one subpopulation with one haplotype, and each red dot around the boxplot represents ETN value of one accession. The gray dotted-lines denote average ETN of one subgroup. Different letters denote significant differences (P < 0.05) based on Duncan’s multiple-range test
Within the QTL ETN9–3 detected in the JX_whole population (Fig. 2a), the OsWRKY74 gene encoding a WRKY transcription factor was identified. OsWRKY74 was involved in tolerance of phosphate starvation, whose overexpression led to a 24% increase in TN (Dai et al. 2015). Six SNPs and one InDel were found in the promoter region, and one non-synonymous SNP was found in CDS (Fig. 4e). There are five haplotypes in total for OsWRKY74, among which Hap3 is the favorable allele for indica subgroup, whereas Hap3 and Hap4 are favorable alleles for nonind subgroup (Fig. 4f). The colocalization analysis above suggested that our GWAS results were reliable because several previously reported TN-associated genes were identified in this study.
Next, we carried out an in silico analysis of the pyramiding effect of the favorable alleles of the above four genes on ETN. Among the nonind subgroup, 34, 42, 78 and 39 accessions harbor Hap5 of OsAAP1, Hap3/6 of DWL2, Hap2 of NAL1, Hap3/4 of WRKY74, respectively. Interestingly, there are 18 accessions harboring favorable alleles of all the four genes (Additional file 5: Fig. S2a), which showed more ETN compared with accessions with favorable allele of only one gene. Pyramiding favorable alleles of four genes led to 60.2% and 56.5% increase in ETN compared with the average ETN of nonind subgroup in Jiangxi and Hainan (Additional file 5: Fig. S2 b, c), respectively.
Identification of Novel ETN-Associated QTLs
We aimed to identify novel ETN-associated QTLs in two ways. One is mining genes homologous to known tillering-related genes, and another is mining genes involved in plant hormone synthesis or signal transduction, including SL, auxin, cytokinin and GA. In total, we identified 25 novel candidate genes that may be related to ETN. Among them, 10 genes are involved in auxin synthesis or signal transduction, three genes participate in cytokinin synthesis, two genes participate in carotenoids synthesis, one gene participates in GA signal transduction (Yamaguchi et al. 2008), two genes encode ammonium transporter, two genes encode peptides/amino acid transporter and five genes encode sugar/glucose transporter. Detailed information of these 25 genes is listed in Additional file 2: Table S2.
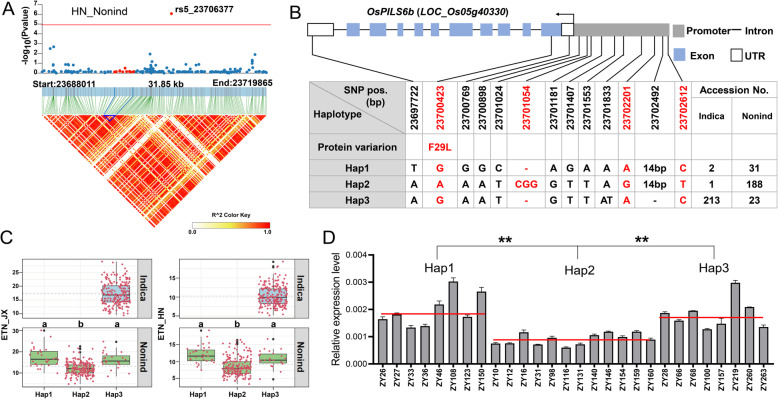
Among the 25 novel candidate genes, we selected the OsPILS6b gene (LOC_Os05g40330) for further analysis, which encodes an auxin efflux carrier protein. Several members of this gene family have been reported to affect rice tillering, including OsPIN2 (Chen et al. 2012), OsPIN5b (Lu et al. 2015a) and OsPIN9 (Hou et al. 2021). LD analysis showed that OsPILS6b located in a relatively high LD region (Fig. 5a). A total of 8 sequence variations were identified in OsPILS6b, including five SNPs and one InDel in the promoter region, one non-synonymous SNP in CDS, and one SNP in 3′-UTR (Fig. 5b). Based on these variations, three haplotypes were identified. The indica subgroup has only one haplotype (Hap3) while the nonind subgroup has all the three haplotypes. Interestingly, most nonind accessions were found in Hap2 and had relatively low ETN (Fig. 5c). Considering the fact that overexpression of OsPIN2 and OsPIN9 have been reported to increase tiller number (Chen et al. 2012; Hou et al. 2021), we detected the OsPILS6b expression levels in accessions with different haplotypes. As expected, accessions with Hap2 had lower expression levels in OsPILS6b than those with Hap1 and Hap3 (Fig. 5d), indicating that the expression level of OsPILS6b is indeed associated with rice tillering.
Fig. 5.
Haplotype and transcription analysis of OsPILS6b. (a) Regional Manhattan plots and LD heatmap of OsPILS6b. (b) Gene structure and SNP information of OsPILS6b. Red characters marked the specific variations for Hap2. The 14-bp deletion in Hap3 is AAAATGGCGGATTG. (c) Haplotype analysis of OsPILS6b. Boxplot described ETN distribution of one subpopulation with one haplotype, and each red dot around the boxplot represents ETN value of one accession. The gray dotted-lines denote average ETN of one subgroup. Different letters denote significant differences (P < 0.05) based on Duncan’s multiple-range test. (d) The relative expression level of OsPILS6b in plants with different haplotypes. Red lines represent the average expression level of accessions with three haplotypes, respectively. “ZYx” is the field number and the detailed information was listed in Additional file 1: Table S1. Data are mean ± SEM, ** means Student’s t-test, P < 0.01
To find out the causative variation for the lower expression in OsPILS6b in accessions with Hap2, we resequenced the OsPILS6b genomic region of the 28 accessions used for RT-qPCR, including 2.5 kb upstream of the start codon ATG. The result showed that besides the variations described above, two SNPs and one 3-bp InDel specific for Hap2 were detected in the promoter region of OsPILS6b (Fig. 5b). We suppose these variations affect the regulation activity of some transcription factors (TF), thus affect the OsPILS6b gene expression. Therefore, we extracted the 100 bp flanking sequence (50 bp upstream and 50 bp downstream) of the three variation and predicted the TF binding sites using the online tool PlantRegMap (https://plantregmap.gao-lab.org/) (Tian et al. 2020). A total of 5, 3 and 261 potential binding sites were located in the flanking sequences of two SNPs and the 3-bp InDel (Additional file 4: Table S4). Notably, it seems that the sequence near the 3-bp InDel is a binding hotspot for multiple transcription factors. Thus we speculate that this InDel is most likely the responsible site for the different expression levels of OsPILS6b among the three halplotypes.
Temporal Expression Pattern of ETN-Associated Genes
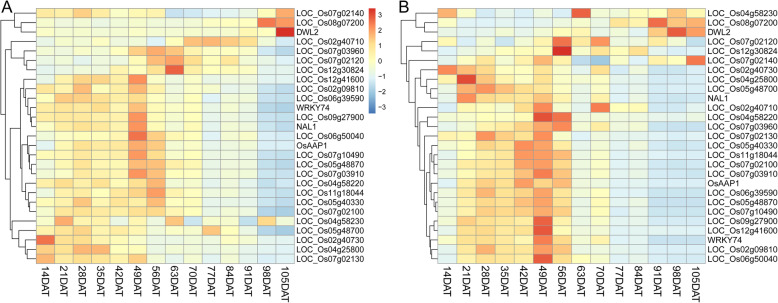
A recent study reported that most tillering-related genes showed a special spatio-temporal expression pattern during the whole growth period. Their expression in root at 00:00 (R0) and 12:00 (R12) are steadily high from 20 days after transplanting (DAT) to 48 DAT and then decrease after 48 DAT (Ma et al. 2020). This pattern may genetically explain the steady transition from tiller development to panicle development. Enlighted by this, we investigated the temporal expression pattern of the ETN-associated genes identified in this study. We did expression clustering analysis using the expression data of root collected at 00:00 (R0) and 12:00 (R12) with weekly interval during the whole growth period from RiceXPro website (http://ricexpro.dna.affrc.go.jp/) (Sato et al. 2011). As expected, among the 27 genes with expression data available in RiceXPro (Additional file 3: Table S3), 17 and 20 genes showed a higher expression from 21 DAT to 49 DAT in R0 and R12, respectively (Fig. 6). This expression pattern coincides with the dynamic change pattern of tiller number during the whole growth period where tiller number increases rapidly from 21 DAT and reaches the peak at 49 DAT (Liu et al. 2018; Liu et al. 2021). The decreased expression of ETN-associated genes after 49 DAT may contribute to the transition from vegetative to reproductive growth of tillers and panicle development on tillers.
Fig. 6.
Temporal expression pattern of ETN-associated genes during the whole growth period. Expression data in root at 00:00 (a) and 12:00 (b) were downloaded from RiceXPro website (https://ricexpro.dna.affrc.go.jp/). The heatmaps represented hierarchical clustering of relative expression levels of 27 candidate genes at different days after transplanting (DAT). The scale for relative expression levels (after normalized by z-score) is denoted by color bars, with red representing the high expression levels, white medium expression and blue low expression
Discussion
GWAS Is a Feasible Way to Mine ETN-Associated QTL
ETN directly determines rice grain yield. Most of the previous studies on ETN were based on classical QTL mapping using biparental populations (Xu and Shen 1991; Wu et al. 1999; Yang et al. 2006; Liu et al. 2010), and few of these QTLs were cloned actually. Traditional QTL mapping can only exploit genetic variations in two parents. However, GWAS can take advantage of extensive variations in numerous natural population (Wang et al. 2020b). Using a panel of 490 rice accessions selected from the 3000 rice genomes project (Wang et al. 2018) and millions of SNPs derived from the same project, we performed GWAS of ETN in two locations. We identified 38 ETN-associated QTLs and found that 1134 genes located in these QTL regions. A recent study on transcriptional regulation of strigolactone signaling in Arabidopsis revealed that genes involved in microtubule function were up-regulated whereas auxin-inducible genes were down-regulated after GR24 treatment for 2 h (Wang et al. 2020a). Among the 1134 genes identified from our GWAS results, three genes are involved in microtubule function. LOC_Os04g58130 (OsKTN80b) encodes a katanin protein showing microtubule-severing activity, LOC_Os05g02670 encodes a kinesin protein involved in microtubule-based movement, and LOC_Os09g27700 encodes a microtubule-associated protein (MAP65/ASE1). Besides, 10 genes associated with auxin synthesis or signal transduction pathways were detected (Additional file 2: Table S2), consistent with the fact that plant hormone synthesis or signal transduction have a significant impact on rice tillering. These results hinted that genes detected from our GWAS results potentially play a role in rice tillering regulation.
Some other identified genes are also involved in plant hormones or homologous to known tillering-related genes. For instance, LOC_Os06g50040 (OsSAUR29) and LOC_Os12g41600 (OsSAUR57), two small auxin-up RNA genes, are homologues of SAUR39 and SAUR27 which have a negative effect on rice tillering (Kant et al. 2009; Ma et al. 2020). LOC_Os07g10490 (ZDS2) encodes a zeta-carotene desaturase involved in the biosynthesis of carotenoids, the precursor of the plant hormone strigolactone. Indeed, genes involved in carotenoids biosynthesis have been reported to regulate rice tillering via the carotenoid-dependent strigolactone biosynthesis, such as the MIT1 gene encoding 15-cis-ζ-carotene isomerase (Z-ISO) (Liu et al. 2020; Zhou et al. 2021; Liu et al. 2021) and the MIT3 gene encoding carotenoid isomerase (CRTISO) (Liu et al. 2018). LOC_Os02g09810 encodes an amino acid transporter-like protein, and four members of this gene family i.e., OsAAP1 (Ji et al. 2020), OsAAP3 (Lu et al. 2018), OsAAP4 (Fang et al. 2021) and OsAAP5 (Wang et al. 2019) have been known to affect tillering.
ETN-Associated QTLs Have Potential for Molecular Breeding
We detected four genes previously reported to be associated with ETN, i.e., OsAAP1, DWL2, NAL1, and OsWRKY74. Haplotype analysis revealed that these genes showed different haplotype patterns in the two subpopulations and different ETN variation among haplotypes. Among the four genes, a favorable allele of NAL1 from japonica has already been used to improve yield in modern indica cultivars (Zhang et al. 2014; Fujita et al. 2013). As to the other three genes, there is no report about their breeding value yet. However, our results showed that Hap5 of OsAAP1, Hap3 and Hap6 of DWL2, Hap3 and Hap4 of WRKY74 are favorable alleles for ETN (Fig. 3 and Fig. 4), and the 18 accessions harboring favorable alleles of all these four genes showed an apparent increase in ETN (Additional file 5: Fig. S2), indicating that pyramiding multiple favorable alleles may contribute to high ETN.
Besides the four known genes, we identified 25 novel genes that may also affect ETN, and the OsPILS6b gene encoding a PIN family member was selected for preliminary validation through RT-qPCR. Accessions with Hap1 and Hap3 generally have higher ETN than accessions with Hap2. Interestingly, the expression levels of OsPILS6b were also higher in accessions with Hap1 and Hap3 than those with Hap2 (Fig. 5). This result is consistent with previous findings that OsPIN2 and OsPIN9, two homologs of OsPILS6b, contribute positively to tillering (Chen et al. 2012; Hou et al. 2021). Moreover, using the public expression data from RiceXpro website, we identified a common spatio-temporal expression pattern for ETN-associated genes, which showed a relatively higher expression from 21 DAT to 49 DAT in root and decreased after that (Fig. 6). This expression pattern coincides with the dynamic change pattern of tiller number during the whole growth period (Liu et al. 2018; Liu et al. 2021). We also analyzed the haplotypes of some other genes, including LOC_Os07g10490 (ZDS2), LOC_Os06g39590 (OsIAA23), LOC_Os06g50040 (OsSAUR29) and LOC_Os12g41600 (OsSAUR57) (Additional file 5: Fig. S3). A total of 5, 9, 11, 8 haplotypes were identified for these four genes, respectively. In addition, these four genes displayed different haplotype patterns between the indica and nonind subpopulation, indicating that some of the ETN-associated genes differentiated after the differentiation of japonica and indica subspecies. These results enlighten us to further mine the favorable alleles in natural resources which can be used for in rice breeding for high ETN.
Conclusions
In this study, we identified 38 ETN-associated QTLs through GWAS using a panel of 490 rice accessions. Four QTLs were colocalized with known genes involved in tillering regulation, and 25 novel genes located in these QTL regions were related to plant hormones or homologous to other ETN-associated genes. The favorable alleles mined with GWAS have potential value in rice molecular breeding for high ETN.
Supplementary Information
Additional file 1: Table S1. Summary of the 490 rice accessions and their ETN phenotypes and information of 28 accessions used for OsPILS6b transcription analysis.
Additional file 2: Table S2. Detailed information of significant SNPs and ETN-associated genes mapped by six GWAS assays.
Additional file 3: Table S3. Expression data of 27 ETN-associated genes.
Additional file 4: Table S4. Potential transcription factors and their binding sites in the selected regions of OsPILS6b promoter.
Additional file 5: Figure S1. Cross-validation plot for population structure and linkage disequilibrium analysis. Figure S2. In silico analysis of the pyramiding effect of favorable alleles of OsAAP1, DWL2, NAL1 and WRKY74. Figure S3. Haplotype analysis of ZDS2, OsIAA23, OsSAUR29 and OsSAUR57.
Acknowledgements
We thank Prof. Zhikang Li and Jianlong Xu (Institute of Crop Sciences, Chinese Academy of Agricultural Sciences) for providing the 490 rice accessions.
Abbreviations
- 3 K RGP
3000 Rice Genomes Project
- ANOVA
Analysis of variance
- AM
Axillary meristem
- CDS
Coding sequence
- EMMAX
Efficient mixed model association expedited
- ETN
Effective tiller number
- GAs
Gibberellins
- GWAS
Genome-wide association study
- HN
Hainan Province
- InDel
Insertion and deletion
- JX
Jiangxi Province
- LD
Linkage disequilibrium
- MAF
Minor allele frequency
- NGS
Next-generation sequencing
- PCA
Principal component analysis
- QTL
Quantitative trait locus
- RT-qPCR
Reverse transcription quantitative-PCR
- SNP
Single nucleotide polymorphism
- SLs
Strigolactones
- TN
Tiller number
- UTR
Untranslated region
Authors’ contributions
XL and HH designed the project. HQ, YC, LL and AK finished the phenotype collection. MR, MH, JZ and JF analyzed the data. MR, MH and XL wrote the manuscript. All authors had read and approved the manuscript.
Funding
This research was supported by National Natural Science Foundation of China (31670279, 31271311), National Major Project for Developing New GM Crops (2016ZX08009–003), and the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences.
Availability of data and materials
All data generated or analyzed in this study are available within the manuscript or its supplementary files or are available from the corresponding authors upon request.
Declarations
Ethics approval and consent to participate
No applicable.
Consent for publication
No applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mengmeng Ren and Minghan Huang contributed equally to this work.
Contributor Information
Mengmeng Ren, Email: ren_mm1991@163.com.
Minghan Huang, Email: huangmh@pku.edu.cn.
Haiyang Qiu, Email: 1179716549@qq.com.
Yan Chun, Email: chunyan1003@gmail.com.
Lu Li, Email: lucaslee1@163.com.
Ashmit Kumar, Email: ashmitkumar0801@gmail.com.
Jingjing Fang, Email: fangjingjing@caas.cn.
Jinfeng Zhao, Email: zhaojinfeng@caas.cn.
Hang He, Email: hehang@pku.edu.cn.
Xueyong Li, Email: lixueyong@caas.cn.
References
- Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov N, Tai S, Wang W, Mansueto L, Palis K, Fuentes RR, Ulat VJ, Chebotarov D, Zhang G, Li Z, Mauleon R, Hamilton RS, McNally KL. SNP-seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 2015;43(D1):D1023–D1027. doi: 10.1093/nar/gku1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 2007;51(6):1019–1029. doi: 10.1111/j.1365-313X.2007.03210.x. [DOI] [PubMed] [Google Scholar]
- Chen YN, Fan XR, Song WJ, Zhang YL, Xu GH. Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol J. 2012;10(2):139–149. doi: 10.1111/j.1467-7652.2011.00637.x. [DOI] [PubMed] [Google Scholar]
- Dai XY, Wang YY, Zhang WH. OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J Exp Bot. 2015;67:947–960. doi: 10.1093/jxb/erv515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong SS, He WM, Ji JJ, Zhang C, Guo Y, Yang TL. LDBlockShow: a fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files. Brief Bioinform. 2020;bbaa:227. doi: 10.1093/bib/bbaa227. [DOI] [PubMed] [Google Scholar]
- Doyle J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Duan JB, Yu H, Yuan K, Liao ZG, Meng XB, Jing YH, Liu GF, Chu JF, Li JY. Strigolactone promotes cytokinin degradation through transcriptional activation of CYTOKININ OXIDASE/DEHYDROGENASE 9 in rice. Proc Natl Acad Sci U S A. 2019;116(28):14319–14324. doi: 10.1073/pnas.1810980116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Ye SW, Tang JY, Bennett MJ, Liang WQ. DWT1/DWL2 act together with OsPIP5K1 to regulate plant uniform growth in rice. New Phytol. 2020;225(3):1234–1246. doi: 10.1111/nph.16216. [DOI] [PubMed] [Google Scholar]
- Fang ZM, Wu BW, Ji YY. The amino acid transporter OsAAP4 contributes to rice tillering and grain yield by regulating neutral amino acid allocation through two splicing variants. Rice (N Y) 2021;14(1):2. doi: 10.1186/s12284-020-00446-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita D, Trijatmiko KR, Tagle AG, Sapasap MV, Koide Y, Sasaki K, Tsakirpaloglou N, Gannaban RB, Nishimura T, Yanagihara S, Fukuta Y, Koshiba T, Slamet-Loedin IH, Ishimaru T, Kobayashi N. NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars. Proc Natl Acad Sci U S A. 2013;110(51):20431–20436. doi: 10.1073/pnas.1310790110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun R, Portais JC, Bouwmeester H, Bécard G, Beveridge CA, Rameau C, Rochange SF. Strigolactone inhibition of shoot branching. Nature. 2008;455(7210):189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- Guo ZL, Liu X, Zhang B, Yuan XJ, Xing YZ, Liu HY, Luo LJ, Chen GX, Xiong LZ. Genetic analyses of lodging resistance and yield provide insights into post-green-revolution breeding in rice. Plant Biotechnol J. 2020;19(4):814–829. doi: 10.1111/pbi.13509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou MM, Luo FF, Wu DX, Zhang XH, Lou MM, Shen DF, Yan M, Mao CZ, Fan XR, Xu GH, Zhang YL. OsPIN9, an auxin efflux carrier, is required for the regulation of rice tiller bud outgrowth by ammonium. New Phytol. 2021;229(2):935–949. doi: 10.1111/nph.16901. [DOI] [PubMed] [Google Scholar]
- Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, Li M, Fan D, Guo Y, Wang A, Wang L, Deng L, Li W, Lu Y, Weng Q, Liu K, Huang T, Zhou T, Jing Y, Li W, Lin Z, Buckler ES, Qian Q, Zhang QF, Li J, Han B. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010;42(11):961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- Ji YY, Huang WT, Wu BW, Fang ZM, Wang XL. The amino acid transporter AAP1 mediates growth and grain yield by regulating neutral amino acid uptake and reallocation in Oryza sativa. J Exp Bot. 2020;71(16):4763–4777. doi: 10.1093/jxb/eraa256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Fang JJ, Lou LM, Zhao JF, Yuan SJ, Yin L, Sun W, Peng LX, Guo BT, Li XY. Characterization of a null allelic mutant of the rice NAL1 gene reveals its role in regulating cell division. PLoS One. 2015;10(2):e0118169. doi: 10.1371/journal.pone.0118169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Liu X, Xiong GS, Liu HH, Chen FL, Wang L, Meng XB, Liu GF, Yu H, Yuan YD, Yi W, Zhao LH, Ma HL, He YZ, Wu ZS, Melcher K, Qian Q, Xu HE, Wang YH, Li JY. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature. 2013;504(7480):401–405. doi: 10.1038/nature12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Wang D, Yan SY, Liu SM, Liu B, Kang HX, Wang GL. Dissection of the genetic architecture of rice tillering using a genome-wide association study. Rice (N Y) 2019;12(1):43. doi: 10.1186/s12284-019-0302-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK. Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Bi YM, Zhu Y, Rothstein SJ. SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol. 2009;151(2):691–701. doi: 10.1104/pp.109.143875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumoto T, Shimada H, Kusano H, She K-C, Iwamoto M, Takano M. Rice monoculm mutation moc2, which inhibits outgrowth of the second tillers, is ascribed to lack of a fructose-1,6-bisphosphatase. Plant Biotechnol. 2013;30(1):47–56. doi: 10.5511/plantbiotechnology.12.1210a. [DOI] [Google Scholar]
- Letunic I, Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Lin B, Wang H, Li XM, Yang FF, Ding XH, Yan JB, Chu ZH. Natural variation in ZmFBL41 confers banded leaf and sheath blight resistance in maize. Nat Genet. 2019;51(10):1540–1548. doi: 10.1038/s41588-019-0503-y. [DOI] [PubMed] [Google Scholar]
- Li XY, Qian Q, Fu ZM, Wang YH, Xiong GS, Zeng DL, Wang XQ, Liu XF, Teng S, Hiroshi F, Yuan M, Luo D, Han B, Li JY. Control of tillering in rice. Nature. 2003;422(6932):618–621. doi: 10.1038/nature01518. [DOI] [PubMed] [Google Scholar]
- Liao ZG, Yu H, Duan JB, Yuan K, Yu CJ, Meng XB, Kou LQ, Chen MJ, Jing YH, Liu GF, Smith SM, Li JY. SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice. Nat Commun. 2019;10(1):2738. doi: 10.1038/s41467-019-10667-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Wang RX, Qian Q, Yan MX, Meng XB, Fu ZM, Yan CY, Jiang B, Su Z, Li JY. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell. 2009;21(5):1512–1525. doi: 10.1105/tpc.109.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GF, Zhu HT, Liu SW, Zeng RZ, Zhang ZM, Li WT, Ding XH, Zhao FM, Zhang GQ. Unconditional and conditional QTL mapping for the developmental behavior of tiller number in rice (Oryza sativa L.) Genetica. 2010;138(8):885–893. doi: 10.1007/s10709-010-9471-y. [DOI] [PubMed] [Google Scholar]
- Liu HJ, Yan JB. Crop genome-wide association study: a harvest of biological relevance. Plant J. 2019;97(1):8–18. doi: 10.1111/tpj.14139. [DOI] [PubMed] [Google Scholar]
- Liu LH, Ren MM, Peng P, Chun Y, Li L, Zhao JF, Fang JJ, Peng LX, Yan JJ, Chu JF, Wang YQ, Yuan SJ, Li XY (2021) MIT1, encoding a 15-cis-ζ-carotene isomerase (Z-ISO), regulates tiller number and stature in rice. J Genet Genomics 10.1016/j.jgg.2020.11.008 [DOI] [PubMed]
- Liu LH, Xie T, Peng P, Qiu HY, Zhao JF, Fang JJ, Patil SB, Wang YQ, Fang S, Chu JF, Yuan S, Zhang WH, Li XY. Mutations in the MIT3 gene encoding a caroteniod isomerase lead to increased tiller number in rice. Plant Sci. 2018;267:1–10. doi: 10.1016/j.plantsci.2017.11.001. [DOI] [PubMed] [Google Scholar]
- Liu WZ, Chao W, Fu YP, Hu GC, Si HM, Li Z, Luan WJ, He ZQ, Sun ZX. Identification and characterization of HTD2: a novel gene negatively regulating tiller bud outgrowth in rice. Planta. 2009;230(4):649–658. doi: 10.1007/s00425-009-0975-6. [DOI] [PubMed] [Google Scholar]
- Liu X, Hu QL, Yan JJ, Sun K, Liang Y, Jia MR, Meng XB, Fang S, Wang YQ, Jing YH, Liu GF, Wu DX, Chu CC, Smith SM, Chu JF, Wang YH, Li JY, Wang B. ζ-Carotene -isomerase suppresses tillering in rice through the coordinated biosynthesis of strigolactone and abscisic acid. Mol Plant. 2020;13(12):1784–1801. doi: 10.1016/j.molp.2020.10.001. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lobell DB, Schlenker W, Costa-Roberts J. Climate trends and global crop production since 1980. Science. 2011;333(6042):616–620. doi: 10.1126/science.1204531. [DOI] [PubMed] [Google Scholar]
- Lu GW, Coneva V, Casaretto JA, Ying S, Mahmood K, Liu F, Nambara E, Bi YM, Rothstein SJ. OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution. Plant J. 2015;83(5):913–925. doi: 10.1111/tpj.12939. [DOI] [PubMed] [Google Scholar]
- Lu K, Wu BW, Wang J, Zhu W, Nie HP, Qian JJ, Huang WT, Fang ZM. Blocking amino acid transporter OsAAP3 improves grain yield by promoting outgrowth buds and increasing tiller number in rice. Plant Biotechnol J. 2018;16(10):1710–1722. doi: 10.1111/pbi.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZF, Shao GN, Xiong JS, Jiao YQ, Wang J, Liu GF, Meng XB, Liang Y, Xiong GS, Wang YH. MONOCULM 3, an ortholog of WUSCHEL in rice, is required for tiller bud formation. J Genet Genomics. 2015;42(2):71–78. doi: 10.1016/j.jgg.2014.12.005. [DOI] [PubMed] [Google Scholar]
- Ma XQ, Li FM, Zhang Q, Wang XQ, Guo HF, Xie JY, Zhu XY, Khan NU, Zhang ZY, Li JJ, Li ZC, Zhang HL. Genetic architecture to cause dynamic change in tiller and panicle numbers revealed by genome-wide association study and transcriptome profile in rice. Plant J. 2020;104(6):1603–1616. doi: 10.1111/tpj.15023. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Kyozuka J. Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell. 2009;21(4):1095–1108. doi: 10.1105/tpc.108.065425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26(7):1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Qian Q, Bu QY, Li SY, Chen Q, Sun JQ, Liang WX, Zhou YH, Chu CC, Li XG, Ren FG, Palme K, Zhao BR, Chen JF, Chen MS, Li CY. Mutation of the Rice Gene, Which Encodes a Novel Protein, Affects Vein Patterning and Polar Auxin Transport. Plant Physiol. 2008;147(4):1947–1959. doi: 10.1104/pp.108.118778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Antonio BA, Namiki N, Takehisa H, Minami H, Kamatsuki K, Sugimoto K, Shimizu Y, Hirochika H, Nagamura Y. RiceXPro: a platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res. 2011;39:D1141–D1148. doi: 10.1093/nar/gkq1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si LZ, Chen JY, Huang XH, Gong H, Luo JH, Hou QQ, Zhou TY, Lu TT, Zhu JJ, Shangguan YY, Chen E, Gong CX, Zhao Q, Jing YF, Zhao Y, Li Y, Cui LL, Fan DL, Lu YQ, Weng QJ, Wang YC, Zhan QL, Liu KY, Wei XH, An K, An G, Han B. OsSPL13 controls grain size in cultivated rice. Nat Genet. 2016;48(4):447–456. doi: 10.1038/ng.3518. [DOI] [PubMed] [Google Scholar]
- Sudhir K, Glen S, Michael L, Christina K, Koichiro T. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi H, Zhang Y, Hattori S, Omae M, Shimizu-Sato S, Oikawa T, Qian Q, Nishimura M, Kitano H, Xie H, Fang XH, Yoshida H, Kyozuka J, Chen F, Sato Y. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell. 2011;23(9):3276–3287. doi: 10.1105/tpc.111.088765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka W, Ohmori Y, Ushijima T, Matsusaka H, Matsushita T, Kumamaru T, Kawano S, Hirano HY. Axillary meristem formation in rice requires the WUSCHEL ortholog TILLERS ABSENT1. Plant Cell. 2015;27(4):1173–1184. doi: 10.1105/tpc.15.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WJ, Ye J, Yao XM, Zhao PZ, Xuan W, Tian YL, Zhang YY, Xu S, An HZ, Chen GM, Yu J, Wu W, Ge WY, Liu XL, Li J, Zhang HZ, Zhao Y, Yang B, Jiang XZ, Peng C, Zhou C, Terzaghi W, Wang CM, Wan JM. Genome-wide associated study identifies NAC42-activated nitrate transporter conferring high nitrogen use efficiency in rice. Nat Commun. 2019;10:1–11. doi: 10.1038/s41467-018-07882-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Yang DC, Meng YQ, Jin J, Gao G. PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 2020;48(D1):D1104–D1113. doi: 10.1093/nar/gkz1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SD (2018) Qqman: an R package for visualizing GWAS results using Q-Q and Manhattan plots. bioRxiv:005165 10.1101/005165
- Wang J, Wu B, Lu K, Wei Q, Qian JJ, Chen YP, Fang ZM. The amino acid permease 5 (OsAAP5) regulates tiller number and grain yield in rice. Plant Physiol. 2019;180(2):1031–1045. doi: 10.1104/pp.19.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang B, Yu H, Guo H, Lin T, Kou L, Wang AQ, Shao N, Ma HY, Xiong GS, Li X, Yang J, Chu JF, Li JY. Transcriptional regulation of strigolactone signalling in Arabidopsis. Nature. 2020;583(7815):277–281. doi: 10.1038/s41586-020-2382-x. [DOI] [PubMed] [Google Scholar]
- Wang Q, Tang JL, Han B, Huang XH. Advances in genome-wide association studies of complex traits in rice. Theor Appl Genet. 2020;133(5):1415–1425. doi: 10.1007/s00122-019-03473-3. [DOI] [PubMed] [Google Scholar]
- Wang WF, Li G, Zhao J, Chu HW, Lin WH, Zhang DB, Wang ZY, Liang WQ. Dwarf Tiller1, a Wuschel-related homeobox transcription factor, is required for tiller growth in rice. PLoS Genet. 2014;10(3):e1004154. doi: 10.1371/journal.pgen.1004154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WS, Mauleon R, Hu ZQ, Chebotarov D, Tai SS, Wu ZC, Li M, Zheng TQ, Fuentes RR, Zhang F, Mansueto L, Copetti D, Sanciangco M, Palis KC, Xu JL, Sun C, Fu BY, Zhang HL, Gao YM, Zhao XQ, Shen F, Cui X, Yu H, Li ZC, Chen ML, Detras J, Zhou YL, Zhang XY, Zhao Y, Kudrna X, Wang CC, Li R, Jia B, Lu JY, He XC, Dong ZT, Xu JB, Li YH, Wang M, Shi JX, Li J, Zhang DB, Lee S, Hu WS, Poliakov A, Dubchak I, Ulat VJ, Borja FN, Mendoza JR, Ali J, Li J, Gao Q, Niu YC, Yue Z, Naredo ME, Talag J, Wang XQ, Li JJ, Fang XD, Yin H, Glaszmann JC, Zhang JW, Li JY, Hamilton RS, Wing RA, Ruan J, Zhang GY, Wei CC, Alexandrov N, McNally KL, Li ZK, Leung H. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature. 2018;557(7703):43–49. doi: 10.1038/s41586-018-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XL, Wang HW, Liu SX, Ferjani A, Li JS, Yan JB, Yang XH, Qin F. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat Genet. 2016;48(10):1233–1241. doi: 10.1038/ng.3636. [DOI] [PubMed] [Google Scholar]
- Wang YH, Li JY. Branching in rice. Curr Opin Plant Biol. 2011;14(1):94–99. doi: 10.1016/j.pbi.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Wickham H (2016) ggplot2: elegant graphics for data analysis. Springer-Verlag New York. ISBN 978-3-319-24277-4
- Wu WR, Li WM, Tang DZ, Lu HR, Worland AJ. Time-related mapping of quantitative trait loci underlying tiller number in rice. Genetics. 1999;151(1):297–303. doi: 10.1093/genetics/151.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing YZ, Zhang QF. Genetic and molecular bases of rice yield. Annu Rev Plant Biol. 2010;61(1):421–442. doi: 10.1146/annurev-arplant-042809-112209. [DOI] [PubMed] [Google Scholar]
- Xu YB, Shen ZT. Diallel analysis of tiller number at different growth stages in rice (Oryza sativa L.) Theor Appl Genet. 1991;83(2):243–249. doi: 10.1007/BF00226258. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. Gibberellin metabolism and its regulation. Annu Rev Plant Biol. 2008;59(1):225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- Yang GH, Xing YZ, Li SQ, Ding JZ, Yue B, Deng K, Li YS, Zhu YH. Molecular dissection of developmental behavior of tiller number and plant height and their relationship in rice (Oryza sativa L.) Hereditas. 2006;143(2006):236–245. doi: 10.1111/j.2006.0018-0661.01959.x. [DOI] [PubMed] [Google Scholar]
- Yano K, Yamamoto E, Aya K, Takeuchi H, Lo PC, Hu L, Yamasaki 3, Shinya Yoshida M, Kitano H, Hirano K, Matsuoka M (2016) Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat Genet 48(8):927-934. 10.1038/ng.3596. [DOI] [PubMed]
- Yang WN, Guo ZL, Huang CL, Duan LF, Chen GX, Jiang N, Fang W, Feng H, Xie WB, Lian XM, Wang GW, Luo QM, Zhang QF, Liu Q, Xiong LZ. Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat Commun. 2014;5(1):5087. doi: 10.1038/ncomms6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuno N, Takamure I, Kidou S, Tokuji Y, Ureshi A, Funabiki A, Ashikaga K, Yamanouchi U, Yano M, Kato K. Rice shoot branching requires an ATP-binding cassette subfamily G protein. New Phytol. 2009;182(1):91–101. doi: 10.1111/j.1469-8137.2008.02724.x. [DOI] [PubMed] [Google Scholar]
- Yuan S, Wang T, Yin L, Zhao J, Wan J, Li X. Cloning and expression of gene responsible for high-tillering dwarf phenotype in Indica rice mutant gsor23. Rice Sci. 2013;20:320–328. doi: 10.1016/S1672-6308(13)60134-1. [DOI] [Google Scholar]
- Zhang C, Dong SS, Xu JY, He WM, Yang TL. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics. 2019;35:1786–1788. doi: 10.1093/bioinformatics/bty875. [DOI] [PubMed] [Google Scholar]
- Zhang GH, Li SY, Wang L, Ye WJ, Zeng DL, Rao YC, Peng YL, Hu J, Yang YL, Xu J, Ren DY, Gao ZY, Zhu L, Dong GJ, Hu XM, Yan MX, Guo LB, Li CY, Qian Q. LSCHL4 from japonica cultivar, which is allelic to NAL1, increases yield of indica super rice 93-11. Mol Plant. 2014;7(8):1350–1364. doi: 10.1093/mp/ssu055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Chen HY, Ren D, Tang HW, Qiu R, Feng JL, Long YM, Niu BX, Chen DP, Zhong TY, Liu YG, Guo JX. Genetic interactions between diverged alleles of Early heading date 1 (Ehd1) and Heading date 3a (Hd3a)/RICE FLOWERING LOCUS T1 (RFT1) control differential heading and contribute to regional adaptation in rice (Oryza sativa) New Phytol. 2015;208(3):936–948. doi: 10.1111/nph.13503. [DOI] [PubMed] [Google Scholar]
- Zhao J, Wang T, Wang M, Liu Y, Yuan S, Gao Y, Yin L, Sun W, Peng L, Zhang W, Wan J, Li X. DWARF3 participates in an SCF complex and associates with DWARF14 to suppress rice shoot branching. Plant Cell Physiol. 2014;55(6):1096–1109. doi: 10.1093/pcp/pcu045. [DOI] [PubMed] [Google Scholar]
- Zhou F, Lin QB, Zhu LH, Ren YL, Zhou KN, Shabek N, Wu FQ, Mao HB, Dong W, Gan L, Ma WW, Gao H, Chen J, Yang C, Wang D, Tan JJ, Zhang X, Guo XP, Wang JL, Jiang L, Liu X, Chen WQ, Chu JF, Yan CY, Ueno K, Ito S, Asami T, Cheng ZJ, Wang J, Lei CL, Zhai HQ, Wu CY, Wang HY, Zheng N, Wan JM. D14–SCF D3-dependent degradation of D53 regulates strigolactone signalling. Nature. 2013;504(7480):406–410. doi: 10.1038/nature12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Yang M, Zhao L, Zhu ZF, Liu FX, Sun HY, Sun CQ, Tan LB. HIGH-TILLERING AND DWARF 12 regulates photosynthesis and plant architecture by affecting carotenoid biosynthesis in rice. J Exp Bot. 2021;72(4):1212–1224. doi: 10.1093/jxb/eraa497. [DOI] [PubMed] [Google Scholar]
- Zou JH, Zhang SY, Zhang WP, Li G, Chen ZX, Zhai WX, Zhao XF, Pan XB, Xie Q, Zhu LH. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 2006;48(5):687–698. doi: 10.1111/j.1365-313X.2006.02916.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Summary of the 490 rice accessions and their ETN phenotypes and information of 28 accessions used for OsPILS6b transcription analysis.
Additional file 2: Table S2. Detailed information of significant SNPs and ETN-associated genes mapped by six GWAS assays.
Additional file 3: Table S3. Expression data of 27 ETN-associated genes.
Additional file 4: Table S4. Potential transcription factors and their binding sites in the selected regions of OsPILS6b promoter.
Additional file 5: Figure S1. Cross-validation plot for population structure and linkage disequilibrium analysis. Figure S2. In silico analysis of the pyramiding effect of favorable alleles of OsAAP1, DWL2, NAL1 and WRKY74. Figure S3. Haplotype analysis of ZDS2, OsIAA23, OsSAUR29 and OsSAUR57.
Data Availability Statement
All data generated or analyzed in this study are available within the manuscript or its supplementary files or are available from the corresponding authors upon request.