Highlights
-
•
• A novel Vcan mouse allele, VcanAA, has ADAMTS protease-resistant versican.
-
•
• VcanAA/AA mice are viable and develop soft tissue-syndactyly (STS)
-
•
• VcanAA/AA STS is rendered more severe in combination with Adamts20Bt/Bt.
-
•
• Mice lacking the versican GAGβ domain, but not the GAGα domain, also have STS.
-
•
• The versican GAGβ proteolytic fragment versikine is necessary for web regression.
Keywords: Extracellular matrix, Metalloprotease, Proteoglycan, ADAMTS, Limb development, Syndactyly
Abstract
Two inherent challenges in the mechanistic interpretation of protease-deficient phenotypes are defining the specific substrate cleavages whose reduction generates the phenotypes and determining whether the phenotypes result from loss of substrate function, substrate accumulation, or loss of a function(s) embodied in the substrate fragments. Hence, recapitulation of a protease-deficient phenotype by a cleavage-resistant substrate would stringently validate the importance of a proteolytic event and clarify the underlying mechanisms. Versican is a large proteoglycan required for development of the circulatory system and proper limb development, and is cleaved by ADAMTS proteases at the Glu441-Ala442 peptide bond located in its alternatively spliced GAGβ domain. Specific ADAMTS protease mutants have impaired interdigit web regression leading to soft tissue syndactyly that is associated with reduced versican proteolysis. Versikine, the N-terminal proteolytic fragment generated by this cleavage, restores interdigit apoptosis in ADAMTS mutant webs. Here, we report a new mouse transgene, VcanAA, with validated mutations in the GAGβ domain that specifically abolish this proteolytic event. VcanAA/AA mice have partially penetrant hindlimb soft tissue syndactyly. However, Adamts20 inactivation in VcanAA/AA mice leads to fully penetrant, more severe syndactyly affecting all limbs, suggesting that ADAMTS20 cleavage of versican at other sites or of other substrates is an additional requirement for web regression. Indeed, immunostaining with a neoepitope antibody against a cleavage site in the versican GAGα domain demonstrated reduced staining in the absence of ADAMTS20. Significantly, mice with deletion of Vcan exon 8, encoding the GAGβ domain, consistently developed soft tissue syndactyly, whereas mice unable to include exon 7, encoding the GAGα domain in Vcan transcripts, consistently had fully separated digits. These findings suggest that versican is cleaved within each GAG-bearing domain during web regression, and affirms that proteolysis in the GAGβ domain, via generation of versikine, has an essential role in interdigital web regression.
Introduction
Extracellular matrix (ECM) proteolysis is necessary for tissue remodeling during mammalian development. For example, branching morphogenesis, cardiac valve leaflet formation, endochondral ossification and interdigital web regression each require precisely timed and spatially controlled tissue remodeling, which was demonstrated by impairment of these processes in mice with engineered mutations in specific proteases [1], [2], [3], [4], [5], [6]. A number of secreted and cell-surface metalloproteases are implicated in ECM remodeling [7]. Because protease function is realized through substrate cleavage, identifying the most impactful cleavages will elucidate potentially important biological pathways. However, since few proteases have a single substrate, an inherent challenge in interpreting protease-deficient phenotypes is determining which of many possible reduced proteolytic cleavages are responsible for an observed phenotype. A corollary to this statement is that the phenotype may result from reduced proteolysis of several substrates, not just a single one, requiring stringent analysis of each substrate to determine the precise contribution of each. The most rigorous validation, indeed a proof, of the importance of cleavage of a substrate would be partial or complete recapitulation of a phenotype observed in a protease-deficient mutant by a cleavage-resistant substrate [8]. However, such experiments are infrequently undertaken because of the high level of risk involved, since many proteases may attack a protein and a protease could cleave the protein at many sites. This risk reduces the likelihood of definitive insights that could be obtained from rendering a single site resistant to proteolysis. Therefore, defining the significance of specific cleavages in substrates has generally not reached the highest levels of experimental rigor. As a result, the functional landscape of proteolysis during embryonic development, physiological processes and diseases is incompletely annotated.
Versican is a large, aggregating chondroitin sulfate (CS) proteoglycan which is a major constituent of ECM, especially provisional ECM [9]. It is essential for survival beyond 10 days of gestation in the mouse and is required specifically for early development of the entire circulatory system, i.e., the heart, blood vessels and blood cells [10], [11]. It forms networks in ECM via two main mechanisms, i.e., by binding hyaluronan through its N-terminal G1-domain and by interacting with numerous ECM components including fibronectin, fibrillins, fibulin-1 and −2 and tenascin-R and -W via its C-terminal G3 domain (reviewed in [12], [13]).CS chains are covalently attached to large, alternatively spliced core protein domains between the G1 and G3 domains, named GAGα (encoded by Vcan exon 7) and GAGβ (encoded by Vcan exon 8) [14]. Alternative splicing of these exons generates distinct versican variants: V0 (containing both GAGα and GAGβ domains), V1 (containing only the GAGβ domain), V2 (containing only the GAGα domain) and V3 (containing neither GAG-bearing domain). Because of its abundance in embryonic tissues and crucial role in embryogenesis, proteolytic turnover of versican has elicited considerable interest [12].
Among their diverse substrates identified in a variety of biological settings [2], [15], several ADAMTS proteases cleave versican at specific peptide bonds, i.e., E441-A442 (in the GAGβ domain, human and mouse versican V1 isoform enumeration) and E405-A406 (in the GAGα domain, human and mouse versican V2/V0 isoform enumeration) [16]. After proteolysis at E441-A442, the resulting new C-terminus, DPEAAE441 (DPEAAE1428 in the V0 isoform), is specifically detected by a neoepitope antibody, anti-DPEAAE [17], which is widely used to demonstrate versican proteolysis in situ, since this epitope is conserved in humans and mice. The corresponding GAGα domain cleavage in human versican could be recognized by another neo-epitope antibody, anti-NIVSFE [16], whose immunogenic sequence is not conserved in the mouse. ADAMTS 1,4,5,9,15 and 20 can digest versican at the GAGβ site [18], [19], [20], [21], [22] and mouse mutants lacking these proteases demonstrated a requirement for versican GAGβ processing during diverse developmental processes (reviewed in [12]). These processes include interdigital web regression (ADAMTS5, ADAMTS9 and ADAMTS20) [4], [23], myocardial compaction (ADAMTS1) [24], cardiac valve sculpting (ADAMTS5) [25], midline facial closure (ADAMTS9 and ADAMTS20) [23], [26], [27], [28], neural tube closure (ADAMTS9 and ADAMTS20) [28], melanoblast colonization of hair follicles (ADAMTS9 and ADAMTS20) [20], [27], [29], skeletal myotube formation (ADAMTS15) [22], vascular development (ADAMTS1, ADAMTS5 and ADAMTS9) [27], [30], [31] and myometrial activation prior to parturition (ADAMTS9) [32]. The N-terminal G1 domain-bearing versican V1 fragment that extends to the GAGβ cleavage site is referred to as versikine [4], whereas the corresponding versican V2 fragment was previously described as glial-hyaluronic acid-binding protein (GHAP) [16]. In contrast to GAGβ processing, GAGα proteolysis has been less extensively studied and no specific function is yet ascribed to GHAP.
The evidence linking ADAMTS proteases to versican processing in interdigit webs, and the importance of versican proteolysis in this context is compelling. Adamts5, Adamts9 and Adamts20 mRNAs are co-expressed at heightened levels in interdigital web mesenchyme just prior to web regression [4]. Moreover, reduced anti-DPEAAE staining of interdigital webs during the period during which regression occurs was evident in combined Adamts5 + Adamts20 mutant mice, which consistently develop soft-tissue syndactyly (STS) and in mice with limb-specific Adamts9 deletion [4], [23]. Impaired interdigital web regression in these mutants was associated with persistence of versican-rich ECM and a subsequent failure of apoptosis [4], [23]. The N-terminal G1 domain-containing fragment (versikine), was found to induce cell death during interdigital web regression in Adamts5 + Adamts20 mutants, and deletion of one Vcan allele in Adamts20bt/bt mice led to more severe STS, suggesting that versican itself, albeit after proteolysis, could be essential for web regression [4]. Taken together, the findings suggest that cooperative versican processing by ADAMTS proteases generates a critical level of versikine to promote apoptosis of interdigital mesenchymal cells; conversely, when versican proteolysis is reduced below this hypothetical critical threshold, interdigit cells may be resistant to BMP-induced apoptosis [4].
Here, we report the generation and analysis of mice with versican resistant to cleavage at the E441-A442 ADAMTS site, using a technically different approach than that recently utilized to generate another cleavage-resistant Vcan allele (designated Vcan(R/R) in the homozygous state) [33]. Our previously reported experiments had demonstrated that substitution of E441 with A (Ala) reduced, but did not eliminate versican cleavage by ADAMTS5 in vitro and had revealed a cryptic ADAMTS5 cleavage site at E438-A439 (variant V1 sequence enumeration) [34]. Replacement of both glutamic acid residues with alanine, i.e., E438 to A + E441 to A eliminated ADAMTS5 processing in this region of the versican core protein [34]. With this preliminary evidence of specific point mutations that could abrogate versican processing, we used homologous recombination in mouse embryonic stem cells to replace E438 and E441 with A and generate transgenic mice with cleavage-resistant versican, an allele designated as VcanAA. Here, we used this new allele to rigorously test the biological relevance of versican proteolysis at these two sites in the context of web regression and along with analysis of web regression in Vcan exon 7- or exon 8-specific mutants, we provide new insights on the role of ADAMTS proteases and versican in this process.
Results
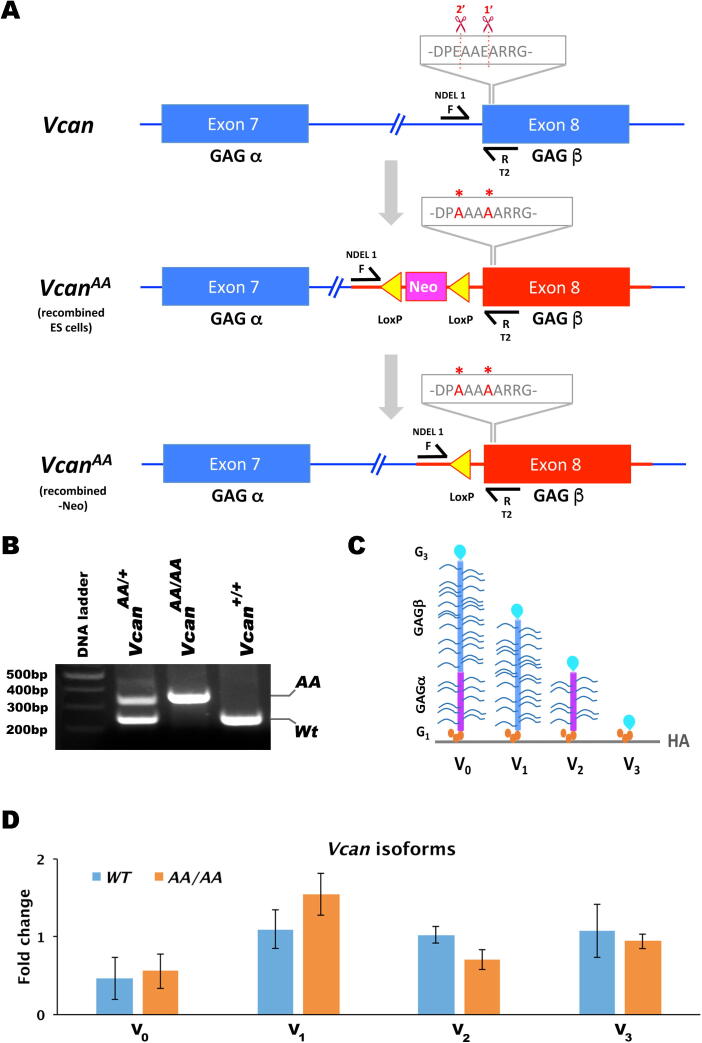
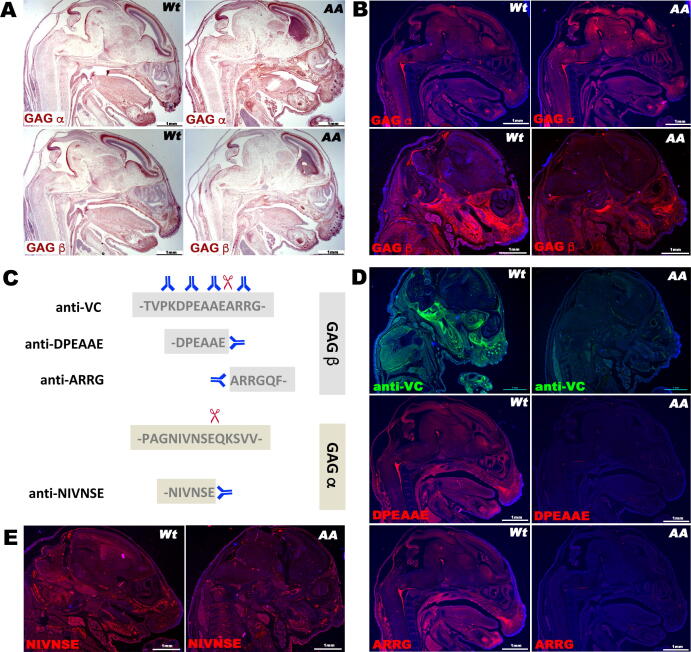
Homologous recombination of a targeting construct bearing the E438 + E441 mutations in ES cells successfully introduced the mutations into the Vcan locus (Fig. 1A, B). Both hemizygous and homozygous mutant mice were viable and fertile. qRT-PCR demonstrated that transcription of the four major Vcan splice isoforms was unaffected (Fig. 1C, D). Furthermore, RNA in situ hybridization demonstrated comparable distribution and intensity of Vcan mRNA using specific exon 7 and exon 8 probes, with each probe showing overlapping expression in wild-type and VcanAA/AA mutant embryos (Fig. 2A). Survival of VcanAA/AA mice indicated absence of the cardiac, vascular and hematopoietic developmental defects seen in Vcanhdf/hdf mutants [11], [35], [36]. Thus, the specific core protein mutations that were introduced did not interfere with these essential functions of versican. Immunostaining using anti-GAGα antibody suggested comparable staining in wild-type and VcanAA/AA embryos (Fig. 2B). Weaker GAGβ immunostaining (with a commercial anti-GAGβ antibody) was seen in VcanAA/AA embryos, since the mutagenized Glu residues occurred within the immunogenic sequence of this antibody (Fig. 2B). Another GAGβ antibody, anti-VC [34], and anti-DPEAAE were unreactive in the mutant mice, which was expected, since the mutated sequence DPAAAA alters epitopes of each antibody (Fig. 2C,D) [34]. Specifically, since the peptide epitope of anti-VC, V436PKDPEAAEARRGQ445 (human versican V1 isoform sequence enumeration; the residues forming the ADAMTS scissile bonds are in bold) is centered on the cleavage sites [34] it was expected to be non-reactive in VcanAA/AA mice, unlike the commercial versican antibody, anti-GAGβ, which is generated to a much longer polypeptide containing the anti-VC epitope and remains weakly reactive (Fig. 2B,D). We therefore used a neo-epitope antibody that detects the new N-terminus, 442ARRGQV, formed by cleavage at the GAGβ site, since this sequence was unaltered by targeted mutagenesis (Fig. 2C). In sections from VcanAA/AA mice, anti-ARRGQV reactivity was also lost, indicating that the engineered mutations abrogated cleavage as intended (Fig. 2D). To investigate cleavage at an ADAMTS processing site previously described in the human versican GAGα domain (NIVSFE405) [16] but uncharacterized in mice, we generated a new neo-epitope antibody, anti-NIVNSE, against the corresponding mouse sequence. The antibody reacted with the immunogen peptide with C-terminal residue E405, which was shown to be crucial for immunoreactivity, but not when E405 was succeeded by another residue (i.e., equivalent to the intact, uncleaved peptide sequence) (Supplemental Fig. 1). Anti-NIVNSE staining was present in both wild-type and mutant embryos (Fig. 2E). To explore whether abrogation of the processing sites would replicate ADAMTS-deficient phenotypes, VcanAA/+ mice were intercrossed at the 3rd (N3) and 9th (N9) generation of outcrosses into the C57BL/6 strain, with the majority of studies being undertaken at N3. Variably penetrant white spotting, a phenotype seen in Adamts20 and Adamts9 mutants [20], [23], [27], [29], was initially seen in a small number of VcanAA/+ and VcanAA/AA mice, but disappeared upon further outcrossing to the C57BL/6 strain. In regard to other ADAMTS protease mutant phenotypes, VcanAA/AA mice lacked cleft palate, a phenotype associated with reduced versican proteolysis in the palatal shelves of mice with combined reduction/inactivation of Adamts9 and Adamts20 [23], [26], [27], [28]. Versican cleavage at the E441-A442 site occurs during ovarian follicular maturation and ovulation, and was previously attributed to ADAMTS1, since Adamts1 mutant mice had impairment of both processes [37], [38], [39]. VcanAA/AA female mice were fertile, suggesting no impact on ovulation and delivered pups without experiencing dystocia, a phenotype reported in mice with smooth muscle-specific conditional inactivation of Adamts9 that was also accompanied by versican accumulation [32].
Fig. 1.
Characterization of a new transgenic allele with cleavage-resistant versican. (A) Transgenic strategy including genotyping strategy, showing Vcan exons 7 and 8 (with their encoded domains) (top), the engineered targeting vector (center) and the recombined allele after homologous recombination (bottom). The scissors indicate two previously identified cleavage sites in the GAGβ domain (sequence shown in single amino acid nomenclature, 1́ and 2́ indicate primary and secondary sites), and the asterisks indicate the introduced Ala replacement of Glu at these sites. The engineered exon 8 is shown in red with the relevant amino acid changes above. Neo indicates the neomycin-resistance cassette used for selection of transfected mouse embryonic stem cell (ESC) clones. Genotyping primers NDEL1F and RT2 are shown. Yellow triangles indicate LoxP sites used to delete the Neo cassette by homologous recombination after identification of correctly targeted embryonic stem cells. (B) Agarose gel electrophoresis of the PCR products obtained from genotyping with primers indicated in A showing the wild type and mutant (AA) bands and their detection in Wt, VcanAA/+ and VcanAA/AA mice. (C) Cartoon depicting the major versican isoforms V0-V3 (not drawn to scale). HA, hyaluronan. G1 and G3 indicate the versican globular domains. (D) The introduced mutations did not affect Vcan transcript levels, shown by RT-qPCR using isoform-specific primers. The arbitrary units are fold change relative to the control (wild type) group. (N = 4 mice from each genotype, error bars = S.E.M.). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 2.
Altered versican immunostaining and proteolysis resulting from the introduced mutations. (A-B) The mutations did not affect distribution of versican transcripts shown in the craniofacial region by RNA in situ hybridization with GAGα (exon 7) or GAGβ (exon 8)-specific RNAscope probes (A, red signal) or protein distribution, shown by immunostaining with domain-specific antibodies (B, red signal), both in E14.5 embryos. (C) Epitopes of cleavage-relevant versican antibodies used in the present work. Scissors indicate the ADAMTS cleavage site. (D) The versican antibodies anti-VC (whose peptide epitope is centered on the cleavage site, unlike anti-versican GAGβ, which is generated to a longer spanning polypeptide) and anti-DPEAAE showed staining in sections from wild-type E14.5 embryos (Wt), but not VcanAA/AA embryos, since the introduced mutations alter the epitopes of these antibodies. In contrast, anti-ARRGQF, which reacts with an N-terminal epitope exposed by cleavage at E441-A442 reacted similarly to anti-DPEAAE in Wt embryos, but also gave no staining in VcanAA/AA embryos, demonstrating lack of cleavage at this site. In contrast, GAGα staining detected by anti-NIVNSE was present in both Wt and VcanAA/AA embryos, since the E405-A406 cleavage site was unmodified. Scale bars in A-E = 1 mm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
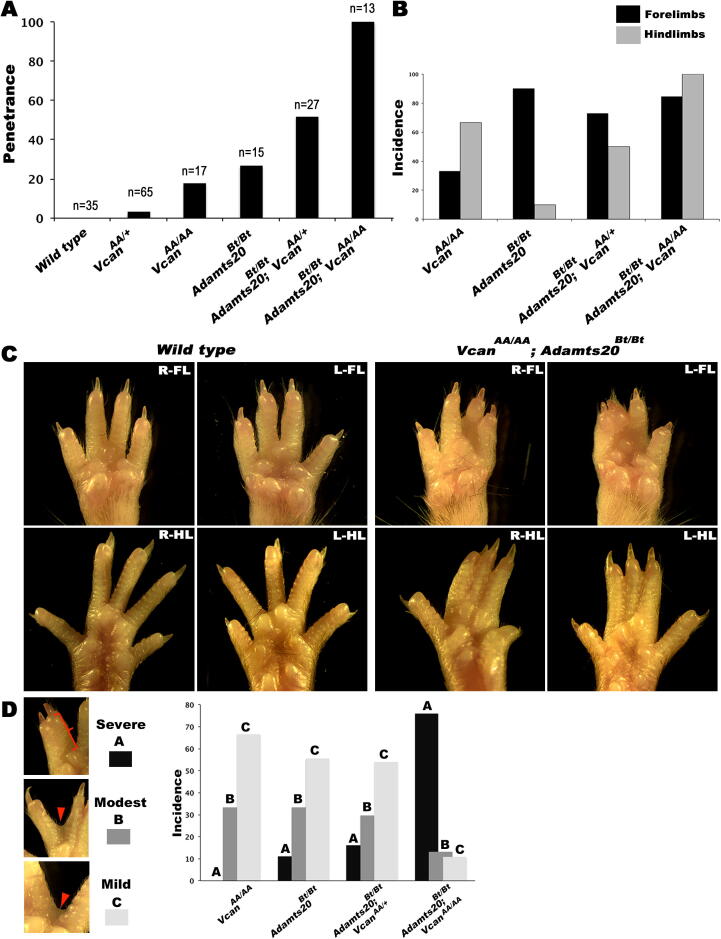
We undertook detailed analysis of interdigital web regression in VcanAA/AA mice, since this is the best characterized of the various defects associated with reduced versican processing. An observed low penetrance of hindlimb STS in VcanAA/+ mice was increased in VcanAA/AA mice, but was nevertheless lower than that seen in Adamts20Bt/Bt mice (Fig. 3A). Introducing one VcanAA/+ allele in Adamts20Bt/Bt mice doubled the penetrance, and STS was fully penetrant in VcanAA/AA;Adamts20Bt/Bt mice, affecting the webs between digits 2 and 3 and digits 3 and 4 (Fig. 3A). Forelimb STS was prevalent in Adamts20Bt/Bt mice, as previously shown [4], whereas hindlimb STS was twice as common as forelimb STS in VcanAA/AA mice (Fig. 3B). Interestingly, VcanAA/AA,Adamts20Bt/Bt mice had roughly equivalent and near-complete penetrance of forelimb and hindlimb STS (Fig. 3B). The grade of STS, i.e., the extent of the syndactylous web along the length of the digit also progressed correspondingly in association with these genotypes (Fig. 3C, D).
Fig. 3.
More severe soft-tissue syndactyly (STS) in mice with cleavage-resistant versican with concurrent Adamts20 deletion. (A) Histogram showing incompletely penetrant STS in VcanAA/+ mice, higher penetrance in VcanAA/AA mice and Adamts20Bt/Bt mice, and complete penetrance in mice carrying both VcanAA and Adamts20 alleles. (B) VcanAA/AA mice had a higher incidence of hindlimb STS, whereas Adamts20Bt/Bt mice had a higher incidence of forelimb STS. VcanAA/AA; Adamts20bt/bt mice had near-complete involvement of both forelimbs and hindlimbs. (C) Illustration of STS severity in forelimbs and hindlimbs. The most severe involvement is observed in VcanAA/AA; Adamts20Bt/Bt mice. Their syndactylous limbs showed consistent involvement of the webs between hindlimb digits 2–3 and 3–4. (D) Classification system for the grade of STS and application to the indicated genotypes. The most severe grade was observed in VcanAA/AA; Adamts20Bt/Bt mice.
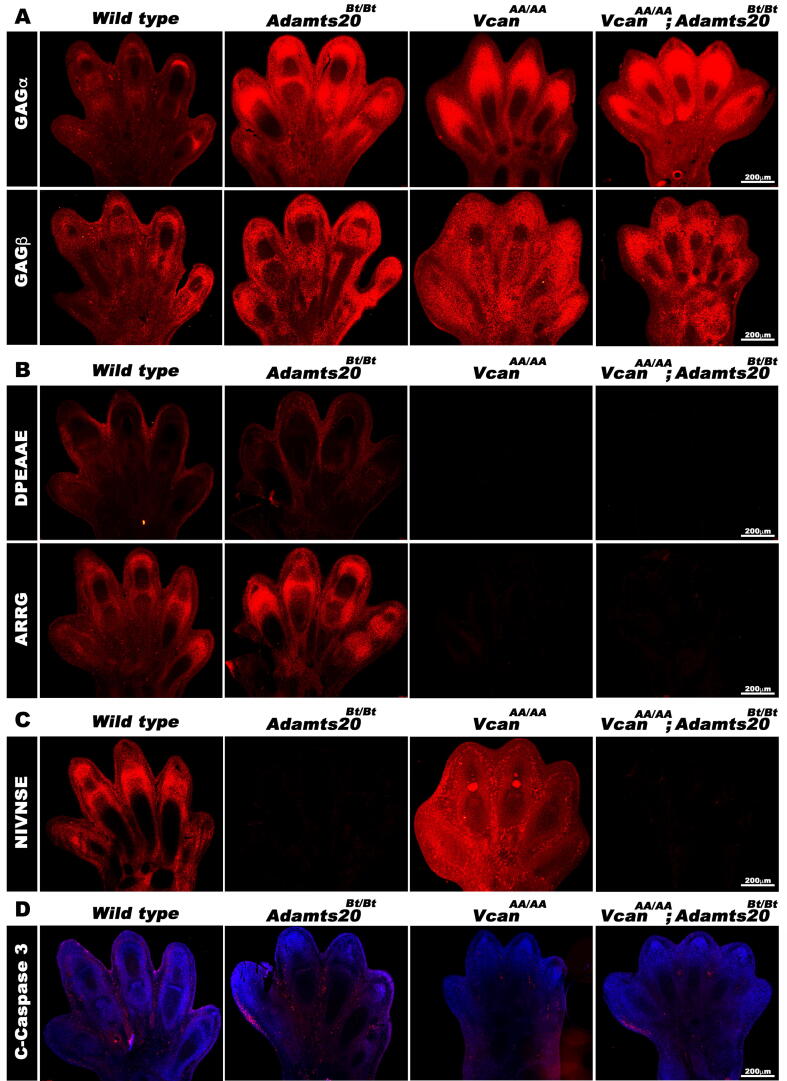
To ascertain the impact of the mutations on versican turnover, we stained for versican and its cleavage products in wild-type, VcanAA/AA, Adamts20Bt/Bt and VcanAA/AA;Adamts20Bt/Bt hindlimb autopods at 13.5 days of gestation. At this developmental stage, web regression is ongoing and incomplete so that it is not possible to anticipate which of the mutant webs will eventually fail to regress. Nevertheless, staining with both GAGα and GAGβ antibodies appeared to be stronger in each of the mutant autopods compared to wild-type autopods (Fig. 4A). As expected, we saw no anti-DPEAAE staining in any mutant with VcanAA/AA alleles (Fig. 4B). DPEAAE staining was detectable in the Adamts20Bt/Bt webs (Fig. 4B), consistent with previous work showing that multiple ADAMTS proteases mediate versican proteolysis at the E441-A442 site [4]. Anti-ARRGQR gave no staining in interdigits from embryos having the VcanAA/AA genotype, demonstrating abrogation of cleavage after mutagenesis of Glu338 and Glu441 (Fig. 4B). Because we observed more intense GAGα staining in Adamts20Bt/Bt mice (Fig. 4A) suggesting that ADAMTS20 might participate in versican cleavage in this core protein domain, we stained autopods of the various genotypes with anti-NIVNSE and found greatly reduced staining in Adamts20Bt/Bt autopods (Fig. 4C). Finally, consistent with the prior finding of reduced apoptosis in syndactylous autopods from ADAMTS mutants, we observed reduced immunostaining for cleaved caspase-3, an apoptosis marker, in VcanAA/AA, Adamts20Bt/Bt and VcanAA/AA;Adamts20Bt/Bt autopods (Fig. 4D).
Fig. 4.
Distribution of versican isoforms (A), cleaved versican (B,C) and apoptotic cells (D) in hind limbs of E13.5 mice of the indicated genotypes. Note increased GAGα and GAGβ staining in each mutant allele shown, whereas anti-DPEAAE, as expected, gives no staining in the presence of mutant versican. Anti-ARRG staining is present in wild-type mice but also in Adamts20Bt/Bt mice, consistent with expression of other ADAMTS proteases that can compensate for the absence of ADAMTS20, contributing to cleavage, but is absent in mice homozygous for the Vcan mutation. Anti-NIVNSE staining is absent in the Adamts20Bt/Bt autopod, but evident in the Vcan mutant, where the GAGα processing site is intact. Reduced cleaved caspase staining is evident in the presence of mutant versican. Scale bars in A-D = 200 μm.
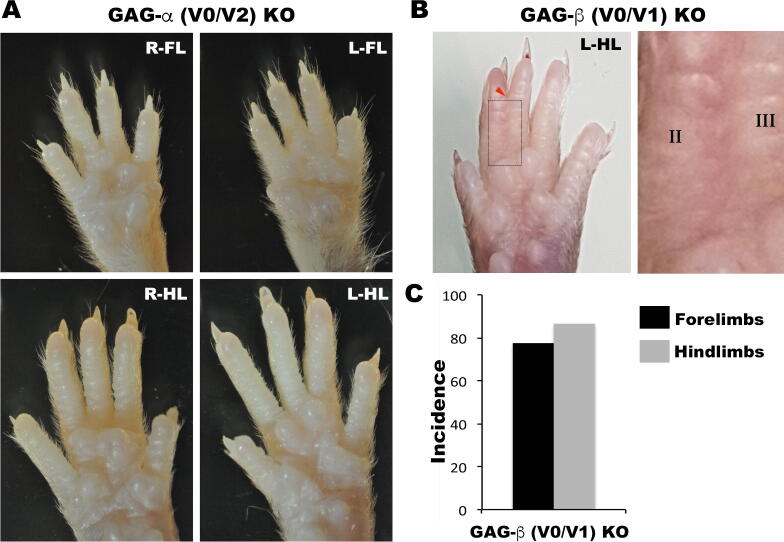
Because prior work had demonstrated that versikine induced cell death in ADAMTS-deficient webs [4], we genetically interrogated the role of versican itself in interdigital web regression by analysis of the limbs of mutants specifically lacking either the GAGα domain [40] or GAGβ domain (Burin Des Roziers and Valleix, manuscript in preparation), which had not been previously done. In contrast to Vcanhdf/hdf mutant mice [11], which lack all versican isoforms, the isoform-specific Vcan mutants survive past birth and into adulthood [40] [and Burin Des Roziers and Valleix, manuscript in preparation]. STS did not occur in the GAGα homozygous mutants which consistently showed fully separated digits, but was seen in all GAGβ homozygous mutants, specifically, 100% (98/98) of GAGβ domain homozygous mutant mice (Fig. 5 A, B). It affected forelimbs in 77% (76/98) of mice and hindlimbs in 86% (85/98) of mice, typically involving the 2–3 and 3–4 interdigit web (Fig. 5 B, C).
Fig. 5.
Soft tissue syndactyly (STS) in Vcan GAGβ but not GAGαknockout mice. (A) STS is not seen in any of the limbs of the Vcan exon 7-lacking mice (GAGα, V0/V2 knockout). (B-C) STS was present with 100% penetrance in Vcan exon 8 mutant mice (GAGβ, V0/V1 knockout), affecting both forelimbs and hindlimbs, and typically the web between digits 2–3 and/or 3–4 (red arrowhead = STS, boxed area in B shown at higher magnification). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Discussion:
In a prior in vitro analysis of versican cleavage by ADAMTS5, we demonstrated that mutation of the P1 and P3 glutamic acid residues (according to cleavage site nomenclature of Schechter and Berger [41]in the versican V1 core protein eliminated versican proteolysis at E441-A442 [34]. On the C-terminal side of the cleavage site, we found that elimination of two proximate GAG chains also reduced processing, likely via removing a putative binding site for the ADAMTS5 ancillary domain [34]. These findings suggested two alternatives for generating a mutant versican core protein that would be cleavage-resistant at the E441-A442 site. We chose to mutate E438 and E441 in the versican core protein in mice since mutation of the residues for GAG chain attachment and elimination of GAG chains could affect the intrinsic properties of versican. In contrast to our approach, Islam et al recently created a cleavage-resistant versican mouse mutant by leaving the DPEAAE sequence intact, but replacing the P1′ to P3′ residues (A442R443R444) by NVY [33]. Despite the intact DPEAAE sequence, Vcan(R/R) mice lacked anti-DPEAAE staining, demonstrating lack of cleavage [33]. Although anti-DPEAAE can no longer be used to detect versican cleavage in VcanAA/AA mutants, lack of anti-ARRGQF reactivity in VcanAA/AA embryos strongly supports elimination of cleavage at the E441-A442 site. Thus, each of these mouse mutants appears to have successfully abrogated embryo-wide proteolysis of versican at the E441-A442 site. Accordingly, hindlimb STS occurred in both VcanAA/AA and Vcan(R/R) mice [33], which carry different mutations engineered using different approaches, i.e., by homologous recombination in ES cells and CRISPR-Cas9 mutagenesis respectively. In contrast to Vcan(R/R) mice [33],VcanAA/AA mutants did not show lethality or other severe phenotypes. A detailed characterization of the VcanAA/AA mice suggested that the inserted mutations did not interfere with Vcan expression or isoform prevalence.
We focused on interdigital web regression during the embryonic period as a classic example of developmental tissue sculpting where versican proteolysis was previously suggested to have a crucial role [4]. The present findings provide novel insights on the role of versican and ADAMTS proteases in web regression. First, occurrence of partially penetrant STS in VcanAA/AA mutants suggests that versican processing at the E441-A442 site contributes to, but does not alone suffice for complete web regression. However, concurrent Adamts20 deletion in VcanAA/AA mutants led to more highly penetrant STS. If ADAMTS20, previously shown to cleave versican at the E441-A442 site [20], cleaved it nowhere else, then its deletion should not have increased STS penetrance and severity. Therefore, we deduced that ADAMTS20 may cleave versican at one or more additional sites in the versican core protein or has one or more additional substrates in the ECM whose proteolysis is necessary for web regression. VcanAA/AA webs that were also lacking ADAMTS20 showed increased staining with a GAGα-specific antibody, indicating higher levels of V0/V2 versican, and we observed reduced anti-NIVNSE staining in these webs. Thus, web regression is associated with cleavage of versican at previously defined sites of ADAMTS proteolysis in both the GAGα and GAGβ domains. Whether ADAMTS20 cleaves versican at additional sites, or cleaves other ECM molecules to enable web regression remains unresolved for now and will be the subject of future studies.
The finding that GAGβ mutant webs but not GAGα mutant webs consistently have defective web regression is consistent with prior work identifying a pro-apoptotic role for versikine, a GAGβ-derived fragment, during web regression [4]. Furthermore, Adamts20Bt/Bt mice haploinsufficient for a Vcan null allele (i.e., Vcanhdf) had more highly penetrant soft-tissue syndactyly than Adamts20Bt/Bt mice, implying that a versican fragment was necessary for web regression [4]. Taken together, these findings strongly support an active role for versican in web regression, not merely as an ADAMTS substrate that is cleared during web regression, but as an essential contributor to this process as the source of versikine. Since the prior experiments that first suggested this possibility [4] used the Vcanhdf mutant allele that lacked all isoforms, the present analysis now confirms that it is the GAGβ domain from which versikine is generated that is crucial. The homozygous GAGβ mutants however have a much higher penetrance of soft-tissue syndactyly than the VcanAA/AA mutants, suggesting that mutating a single cleavage site may not be sufficient to prevent versican proteolysis and that other cleavage sites may exist in the versican core protein that could generate versikine-like activity. This possibility is potentially supported by the higher penetrance resulting from combining the VcanAA/AA and Adamts20Bt/Bt mutants. The data we present also suggests that although GAGα cleavage occurs during web regression, it is not causally involved in the process. However, it is not possible to precisely assess the relative contribution of versikine activity vs versican clearance during web regression. GAGβ proteolysis is undoubtedly essential, and perhaps even an obligate initiating event; however, which regions or motifs of versikine are critical and their underlying mechanisms are undefined. That the GAGα mutant did not develop syndactyly suggests that versikine activity may not reside in the G1 domain, which is common to both versikine and GHAP, but it could reside in the N-terminal segment of the GAGβ domain that constitutes the C-terminal region of versikine.
Mutagenesis of substrates to ascertain the significance of site-specific proteolytic cleavages has been previously used in only a few instances. For example, mice expressing collagen with an introduced mutation that abrogates the classic collagen I cleavage site (Col1a1 (r/r) mice) provided vital information about the role of collagen turnover in bone, wound healing, atherosclerosis and post-partum uterine remodeling [42], [43], [44], [45], [46], [47], [48]. Mice with a mutation that rendered aggrecan resistant to the action of ADAMTS proteases at the E373- A374 site were protected from surgically induced osteoarthritis and inflammatory arthritis, indicating that ADAMTS proteases were major instruments of articular cartilage breakdown in arthritis [49]. Mice with cleavage-resistant reelin have demonstrated the importance of reelin processing by ADAMTS2 and ADAMTS3 in cerebral cortical and hippocampal development [50].
Genetic combination of cleavage-resistant substrate and protease mutants as reported here is rarely used, but can offer new insights. Conceptually, if a protease (e.g., ADAMTS20) has only one substrate (e.g., versican), which it cleaves at a single site (e.g., E441-A442), then genetically combining the cleavage-site mutant and protease mutant should not worsen the phenotype of the cleavage-site mutant. In the only other instance of a similar mutant inter-cross we are aware of, a combination of the Col1a1 (r/r) mouse mutant and Mmp2-/- mice led to more severe phenotypes than either transgene alone, highlighting collagen I cleavage at an alternative site by MMP2 [51]. In this regard, we are currently investigating other sites of ADAMTS proteolysis in the versican core protein.
In the future, VcanAA mutant mice can be investigated more thoroughly for additional morphogenetic roles, such as in the heart, craniofacial development and neural development, or in adult physiological and disease roles. Vcan(R/R) mice demonstrated accelerated wound healing attributed in part to greater myofibroblast activation [52], consistent with prior work showing that pericellular versican accumulation in dermal fibroblasts, achieved either by genetic inactivation of Adamts5 or versican V1 overexpression led to enhanced TGFβ signaling and fibroblast-to-myofibroblast transition [18], [53]. Furthermore, combining the VcanAA mutant with other ADAMTS mouse mutants, such as was done here using Adamts20Bt, may well provide additional insights on the role of versican and ADAMTS processing of versican in the future.
Methods
Targeted mutagenesis of Vcan. The Ensembl transcript Vcan-001 ENSMUST00000109546 (gene ENSMUSG00000021614) was used as the reference for design of a construct targeting the single Vcan locus on chromosome 13. Exon 8 of the 15 Vcan exons generating this transcript encodes the entire GAGβ domain. A ~ 9.2 kb region was subcloned from a C57BL/6 BAC clone (RP23-265 J12) into plasmid vector pSP72 (Promega, Madison, WI), containing an ampicillin selection cassette to construct the targeting vector. The point mutations were engineered by PCR mutagenesis. The engineered mutations in exon 8 altered the sequence from 5́GAAGCTGCAGAA3́ (translated as E438AAE441) to 5́GCAGCTGCAGCA3́ (translated as A438AAA441) and were located 269 nt downstream of the 5′ end of exon 8. The PCR fragment carrying the mutation was then inserted into the vector using a homologous recombination-based method. The targeting vector contained a 5′-homology arm of 3.0 kb juxtaposed to a FRT-LoxP-flanked self-excising neomycin resistance cassette (IV-UBS-FRT-LoxP flanked Neomycin cassette). This cassette was inserted 203 bp upstream of the intron 7-exon 8 splice site. The 3′ homology arm extended 5.7 kb downstream of the introduced mutations. The total size of the targeting construct (including pSP72 vector backbone, Neo cassette and a Diphtheria toxin cassette for negative selection) was 16.2 kb. The targeting vector construction was confirmed by restriction mapping and Sanger sequencing. 10 μg of the targeting vector linearized using NotI was electroporated into HF4 (129/SvEv × C57BL/6) (FLP hybrid) embryonic stem cells (ESC). After selection with G418, surviving ESC clones were expanded for PCR analysis to identify recombinant ESC. Neo was excised by FlpE recombinase during ESC clone expansion leaving behind a 118 nt FRT-loxP tandem site footprint. 4 positive clones identified as positive by Southern blot and PCR screening were selected for further expansion. For Southern blot, genomic DNA was digested with NheI, electrophoresed on a 0.8% gel and hybridized to a 398 bp probe external to the 5′ homology arm as shown. DNA from the HF4 strain was used as a wild-type control. Real-time quantitative PCR showed a single copy of the transgenic vector integrated in ES cells.
Targeted ESC from clone 151 were microinjected into C57BL/6 blastocysts. Resulting chimeras with a high percentage agouti coat color were mated to C57BL/6 wt mice to generate germline Neo deleted mice. 4 heterozygous mice (3 female,1 male) from one high percentage chimera derived from clone 151 were intercrossed to determine the effect of the transgene. Deletion of Neo and FlpE and the presence of the inserted mutations in these mice were confirmed by Sanger sequencing of PCR-amplified genomic DNA using appropriate primers. Outbreeding to the C57Bl/6 strain was undertaken for a further nine generations and mice were analyzed after the third and ninth intercross for the impact of the transgene on interdigital web regression. Mice were carefully examined for the incidence, extent and severity of STS as previously described [4]. Penetrance of STS was defined as a persistent web of any severity in at least one interdigit of a mouse [4].
Other mouse transgenes: The splice variant-specific versican GAGα mutant mouse strain Vcan(tm1Zim) carries a translational stop codon preceded by an ER-retention signal in the GAGα-encoding exon 7. In consequence, versican isoforms V0 and V2 are absent in homozygous animals, while the isoforms V1 and V3 are normally expressed [40]. Mice lacking the GAGβ domain were generated by deleting the entire Vcan exon 8 by CRISPR-Cas 9 gene editing and will be described elsewhere (Burin Des Roziers, C. and Valleix,S., manuscript in preparation). qPCR analysis of the different Vcan transcript isoforms coupled with RNASeq analysis, and immunohistochemistry analysis of GAGβ homozygous mutant tissue has shown an absence of the V0/V1 transcript isoforms, a marked increase in V2/V3 transcript isoforms and absence of GAGβ immunostaining (Burin Des Roziers, C. and Valleix,S., manuscript in preparation).
Antibodies, ELISA and Western blotting. The generation of anti-442ARRGQF, to the new N-terminus generated by cleavage at E441-A442 was previously described [54]. In accordance with well-characterized methods for generation of neo-epitope antibodies [55], rabbits were immunized with the peptide immunogen CGGNIVNSE405 covalently coupled to keyhole limpet hemocyanin. Immune sera were purified by affinity chromatography using the immobilized peptide immunogen as the ligand and tested by ELISA against the same peptide. To determine the specificity of anti-NIVNSE for the neoepitope but not the bridging (intact) peptide sequence and for determining the critical requirements for antibody binding, we utilized ELISA against the immunogen peptides and a number of variant peptides. Peptides (4 μg/ml) were adsorbed to Nunc MaxiSorb plates and reacted with increasing dilutions of the respective antibodies. Anti-DPEAAE441, anti-GAGα and anti-VC were previously described [16], [17], [34].
RNA In situ hybridization and immunofluorescence: Mouse embryos were obtained at 13.5 and 14.5 days of gestation (E13.5 and E14.5) and fixed in 4% paraformaldehyde prior to paraffin-embedding. 7 μm thick sections were used for RNA in situ hybridization utilizing RNAscope technology (Advanced Cell Diagnostics) with specific Vcan probes for exon 7 and exon 8 [10], [28] as previously described [56]. Sections were used for antibody staining by an indirect immunofluorescence method after antigen retrieval, essentially as previously described [10], [28] using anti-cleaved caspase-3 (Asp175), (1:100, Cell Signaling, 9661), versican anti-GAGβ (catalog no. AB1033, MilliporeSigma), anti-GAGα (catalog no. AB1032, MilliporeSigma), anti-VC [34], anti-42ARRGQF [54] as well as anti-NIVNSE, which is described above. DAPI or hematoxylin were used as the counterstain as appropriate. Sections were photographed on an Olympus BX51 upright microscope (Olympus, Center Valley, PA) using a Leica DFC7000T camera and Leica Application Suite v4.6 imaging software (both from Leica, Wetzlar, Germany).
Quantitative real time RT-PCR (qRT-PCR) analysis: RNA was extracted from 23 day old wild type and VcanAA homozygous mouse tissues using TRIzol reagent (ThermoFisher catalog no. 15596026). Lung mRNA was used for Vcan V0, V1 and V3 qRT-PCR analysis and brain mRNA for Vcan V2 qRT-PCR analysis. 2 μg of RNA from each sample was used for cDNA synthesis using the high-capacity cDNA synthesis kit (Applied Biosystems, catalog no. 4368814). qRT-PCR was carried out using a CFX96 Touch Real-Time PCR detection system (BIO-RAD Laboratories) with the Bullseye EvaGreen qPCR mix (MIDSCI, catalog no. BEQPCR-S). 18 s ribosomal mRNA levels were used for normalization and relative expression of genes was calculated using the ΔΔCt quantification method. An unpaired, two-tailed Student t-test was used to determine statistical significance. The primer pairs used were: V0 primers, forward 5′ TTCACAGAACGCCACCCTTGAGTCC 3′ and reverse 5′ CTAGCTTCTGCAGCTTCCGGGTCC 3′; V1 primers, forward 5′ GCAGCTTGGAGAAATGGCTTTGACC 3′ and reverse 5′ CTAGCTTCTGCAGCTTCCGGGTCC 3′; V2 primers forward 5′ TCCTGGAGAATCTGTAACACAGCACCC 3′ and reverse 5′ CTCGGTAGGATAACAGGTGCCTCCG 3′; V3 primers forward 5′ ACTTCAGGCAGCTTGGAGAAATGGC 3′ and reverse 5′ ACTGGTCTCCGCTGTATCCAGGTGC 3′; 18s primers, forward 5′ TTGACGGAAGGGCACCACCAG 3′ , reverse 5′ GCACCACCACCCACGGAATCG 3′.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Institutes of Health Award HL107147 (NHLBI Program of Excellence in Glycoscience; to S.S.A.), by the Allen Distinguished Investigator Program, through support made by The Paul G. Allen Frontiers Group and the American Heart Association (to S.S.A.), the Mark Lauer Pediatric Research Grant (to S.N.) and the Swedish Heart-Lung Foundation (to K.T.L).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mbplus.2021.100064.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Bonnans C., Chou J., Werb Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014;15(12):786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dubail J., Apte S.S. Insights on ADAMTS proteases and ADAMTS-like proteins from mammalian genetics. Matrix Biol. 2015;44–46:24–37. doi: 10.1016/j.matbio.2015.03.001. PubMed PMID: 25770910. [DOI] [PubMed] [Google Scholar]
- 3.Kern C.B., Twal W.O., Mjaatvedt C.H., Fairey S.E., Toole B.P., Iruela-Arispe M.L., Argraves W.S. Proteolytic cleavage of versican during cardiac cushion morphogenesis. Dev. Dyn. 2006;235(8):2238–2247. doi: 10.1002/dvdy.20838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D.R. McCulloch, C.M. Nelson, L.J. Dixon, D.L. Silver, J.D. Wylie, V. Lindner, T. Sasaki, M.A. Cooley, W.S. Argraves, S.S. Apte, ADAMTS metalloproteases generate active versican fragments that regulate interdigital web regression. Dev Cell. 2009, 17(5):687-98. Epub 2009/11/20. S1534-5807(09)00390-6 [pii] 10.1016/j.devcel.2009.09.008. PubMed PMID: 19922873; PMCID: 2780442. [DOI] [PMC free article] [PubMed]
- 5.Oblander S.A., Zhou Z., Galvez B.G., Starcher B., Shannon J.M., Durbeej M., Arroyo A.G., Tryggvason K., Apte S.S. Distinctive functions of membrane type 1 matrix-metalloprotease (MT1-MMP or MMP-14) in lung and submandibular gland development are independent of its role in pro-MMP-2 activation. Dev. Biol. 2005;277(1):255–269. doi: 10.1016/j.ydbio.2004.09.033. PubMed PMID: 15572153. [DOI] [PubMed] [Google Scholar]
- 6.Vu T.H., Shipley J.M., Bergers G., Berger J.E., Helms J.A., Hanahan D., Shapiro S.D., Senior R.M., Werb Z. MMP-9/gelatinase B is a key regulator of growth plate angiogenesis and apoptosis of hypertrophic chondrocytes. Cell. 1998;93(3):411–422. doi: 10.1016/s0092-8674(00)81169-1. PubMed PMID: 9590175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Apte S.S., Parks W.C. Metalloproteinases: A parade of functions in matrix biology and an outlook for the future. Matrix Biol. 2015;44–46:1–6. doi: 10.1016/j.matbio.2015.04.005. PubMed PMID: 25916966. [DOI] [PubMed] [Google Scholar]
- 8.S.S. Apte, ADAMTS Proteins: Concepts, Challenges, and Prospects. Methods Mol Biol. 2020, 2043, 1-12. Epub 2019/08/30. 10.1007/978-1-4939-9698-8_1. PubMed PMID: 31463898. [DOI] [PubMed]
- 9.T.N. Wight, Provisional matrix: A role for versican and hyaluronan. Matrix Biol. 2017, 60-61, 38-56. Epub 2016/12/10. 10.1016/j.matbio.2016.12.001. PubMed PMID: 27932299; PMCID: PMC5438907. [DOI] [PMC free article] [PubMed]
- 10.Nandadasa S., O’Donnell A., Midura R.J., Apte S.S. The versican-hyaluronan complex provides an essential extracellular matrix niche for Flk1+ precursors in vasculogenesis and primitive hematopoiesis. bioRxiv. 2019;753418 doi: 10.1101/753418. [DOI] [Google Scholar]
- 11.Yamamura H., Zhang M., Markwald R.R., Mjaatvedt C.H. A heart segmental defect in the anterior-posterior axis of a transgenic mutant mouse. Dev. Biol. 1997;186(1):58–72. doi: 10.1006/dbio.1997.8559. PubMed PMID: 9188753. [DOI] [PubMed] [Google Scholar]
- 12.Nandadasa S., Foulcer S., Apte S.S. The multiple, complex roles of versican and its proteolytic turnover by ADAMTS proteases during embryogenesis. Matrix Biol. 2014;35:34–41. doi: 10.1016/j.matbio.2014.01.005. PubMed PMID: 24444773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Y.J. Wu, D.P. La Pierre, J. Wu, A.J. Yee, B.B. Yang, The interaction of versican with its binding partners. Cell Res. 2005, 15(7), 483-94. Epub 2005/07/28. 10.1038/sj.cr.7290318. PubMed PMID: 16045811. [DOI] [PubMed]
- 14.Dours-Zimmermann M.T., Zimmermann D.R. A novel glycosaminoglycan attachment domain identified in two alternative splice variants of human versican. J. Biol. Chem. 1994;269(52):32992–32998. PubMed PMID: 7806529. [PubMed] [Google Scholar]
- 15.Mead T.J., Apte S.S. ADAMTS proteins in human disorders. Matrix Biol. 2018;71–72:225–239. doi: 10.1016/j.matbio.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westling J., Gottschall P.E., Thompson V.P., Cockburn A., Perides G., Zimmermann D.R., Sandy J.D. ADAMTS4 (aggrecanase-1) cleaves human brain versican V2 at Glu405-Gln406 to generate glial hyaluronate binding protein. Biochem. J. 2004;377(Pt 3):787–795. doi: 10.1042/BJ20030896. PubMed PMID: 14561220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandy J.D., Westling J., Kenagy R.D., Iruela-Arispe M.L., Verscharen C., Rodriguez-Mazaneque J.C., Zimmermann D.R., Lemire J.M., Fischer J.W., Wight T.N., Clowes A.W. Versican V1 proteolysis in human aorta in vivo occurs at the Glu441- Ala442 bond, a site that is cleaved by recombinant ADAMTS-1 and ADAMTS- 4. J. Biol. Chem. 2001;276(16):13372–13378. doi: 10.1074/jbc.M009737200. PubMed PMID: 11278559. [DOI] [PubMed] [Google Scholar]
- 18.N. Hattori, D.A. Carrino, M.E. Lauer, A. Vasanji, J.D. Wylie, C.M. Nelson, S.S. Apte, Pericellular versican regulates the fibroblast-myofibroblast transition: a role for ADAMTS5 protease-mediated proteolysis. J. Biol. Chem. 2011, 286(39), 34298-310. Epub 2011/08/11. M111.254938 [pii 10.1074/jbc.M111.254938. PubMed PMID: 21828051; PMCID: 3190794 [DOI] [PMC free article] [PubMed]
- 19.J.M. Longpre, D.R. McCulloch, B.H. Koo, J.P. Alexander, S.S. Apte, R. Leduc, Characterization of proADAMTS5 processing by proprotein convertases. Int J Biochem Cell Biol. 2009, 41(5), 1116-26. Epub 2008/11/11. S1357-2725(08)00414-7 [pii] 10.1016/j.biocel.2008.10.008. PubMed PMID: 18992360. [DOI] [PubMed]
- 20.Silver D.L., Hou L., Somerville R., Young M.E., Apte S.S., Pavan W.J., Barsh G. The secreted metalloprotease ADAMTS20 is required for melanoblast survival. PLoS Genet. 2008;4(2):e1000003. doi: 10.1371/journal.pgen.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Somerville R.P., Longpre J.M., Jungers K.A., Engle J.M., Ross M., Evanko S., Wight T.N., Leduc R., Apte S.S. Characterization of ADAMTS-9 and ADAMTS-20 as a distinct ADAMTS subfamily related to Caenorhabditis elegans GON-1. J. Biol. Chem. 2003;278(11):9503–9513. doi: 10.1074/jbc.M211009200. PubMed PMID: 12514189. [DOI] [PubMed] [Google Scholar]
- 22.N. Stupka, C. Kintakas, J.D. White, F.W. Fraser, M. Hanciu, N. Aramaki-Hattori, S. Martin, C. Coles, F. Collier, A.C. Ward, S.S. Apte, D.R. McCulloch, Versican processing by a disintegrin-like and metalloproteinase domain with thrombospondin-1 repeats proteinases-5 and -15 facilitates myoblast fusion. Journal of Biological Chemistry. 2013, 288(3), 1907-17. Epub 2012/12/13. 10.1074/jbc.M112.429647. PubMed PMID: 23233679; PMCID: 3548499. [DOI] [PMC free article] [PubMed]
- 23.Dubail J., Aramaki-Hattori N., Bader H.L., Nelson C.M., Katebi N., Matuska B., Olsen B.R., Apte S.S. A new Adamts9 conditional mouse allele identifies its non-redundant role in interdigital web regression. Genesis. 2014;52(7):702–712. doi: 10.1002/dvg.v52.710.1002/dvg.22784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stankunas K., Hang C.T., Tsun Z.Y., Chen H., Lee N.V., Wu J.I., Shang C., Bayle J.H., Shou W., Iruela-Arispe M.L., Chang C.P. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev. Cell. 2008;14(2):298–311. doi: 10.1016/j.devcel.2007.11.018. PubMed PMID: 18267097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.L.E. Dupuis, D.R. McCulloch, J.D. McGarity, A. Bahan, A. Wessels, D. Weber, A.M. Diminich, C.M. Nelson, S.S. Apte, C.B. Kern, Altered versican cleavage in ADAMTS5 deficient mice; a novel etiology of myxomatous valve disease. Dev Biol. 2011, 357(1), 152-64. Epub 2011/07/14. 10.1016/j.ydbio.2011.06.041. PubMed PMID: 21749862. [DOI] [PMC free article] [PubMed]
- 26.Enomoto H., Nelson C., Somerville R.P.T., Mielke K., Dixon L., Powell K., Apte S.S. Cooperation of two ADAMTS metalloproteases in closure of the mouse palate identifies a requirement for versican proteolysis in regulating palatal mesenchyme proliferation. Development. 2010;137:4029–4038. doi: 10.1242/dev.050591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nandadasa S., Nelson C.M., Apte S.S. ADAMTS9-Mediated Extracellular Matrix Dynamics Regulates Umbilical Cord Vascular Smooth Muscle Differentiation and Rotation. Cell Rep. 2015;11(10):1519–1528. doi: 10.1016/j.celrep.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.S. Nandadasa, C.M. Kraft, L.W. Wang, A. O'Donnell, R. Patel, H.Y. Gee, K. Grobe, T.C. Cox, F. Hildebrandt, S.S. Apte, Secreted metalloproteases ADAMTS9 and ADAMTS20 have a non-canonical role in ciliary vesicle growth during ciliogenesis. Nat Commun. 2019, 10(1), 953. Epub 2019/03/01. 10.1038/s41467-019-08520-7. PubMed PMID: 30814516; PMCID: PMC6393521. [DOI] [PMC free article] [PubMed]
- 29.Rao C., Foernzler D., Loftus S.K., Liu S., McPherson J.D., Jungers K.A., Apte S.S., Pavan W.J., Beier D.R. A defect in a novel ADAMTS family member is the cause of the belted white-spotting mutation. Development. 2003;130(19):4665–4672. doi: 10.1242/dev.00668. PubMed PMID: 12925592. [DOI] [PubMed] [Google Scholar]
- 30.C.C. Hong, A.T. Tang, M.R. Detter, J.P. Choi, R. Wang, X. Yang, A.A. Guerrero, C.F. Wittig, N. Hobson, R. Girard, R. Lightle, T. Moore, R. Shenkar, S.P. Polster, L.M. Goddard, A.A. Ren, N.A. Leu, S. Sterling, J. Yang, L. Li, M. Chen, P. Mericko-Ishizuka, L.E. Dow, H. Watanabe, M. Schwaninger, W. Min, D.A. Marchuk, X. Zheng, I.A. Awad, M.L. Kahn, Cerebral cavernous malformations are driven by ADAMTS5 proteolysis of versican. J. Exp. Med. 2020, 217(10). Epub 2020/07/11. 10.1084/jem.20200140. PubMed PMID: 32648916; PMCID: PMC7537394 work. No other disclosures were reported. [DOI] [PMC free article] [PubMed]
- 31.S. Nandadasa, J.M. Szafron, V. Pathak, S.-I. Murtada, C.M. Kraft, A. O’Donnell, C. Norvik, C. Hughes, B. Caterson, M.S. Domowicz, N.B. Schwartz, K. Tran-Lundmark, M. Veigl, D. Sedwick, E.H. Philipson, J.D. Humphrey, S.S. Apte, Vascular dimorphism ensured by regulated proteoglycan dynamics favors rapid umbilical artery closure at birth. bioRxiv. 2020, 2020.07.02.184978. 10.1101/2020.07.02.184978. [DOI] [PMC free article] [PubMed]
- 32.Mead T.J., Du Y., Nelson C.M., Gueye N.A., Drazba J., Dancevic C.M., Vankemmelbeke M., Buttle D.J., Apte S.S. ADAMTS9-Regulated Pericellular Matrix Dynamics Governs Focal Adhesion-Dependent Smooth Muscle Differentiation. Cell Rep. 2018;23(2):485–498. doi: 10.1016/j.celrep.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.S. Islam, K. Chuensirikulchai, S. Khummuang, T. Keratibumrungpong, P. Kongtawelert, W. Kasinrerk, S. Hatano, A. Nagamachi, H. Honda, H. Watanabe, Accumulation of versican facilitates wound healing: implication of its initial ADAMTS-cleavage site. Matrix Biol. 2019. Epub 2019/11/02. 10.1016/j.matbio.2019.10.006. PubMed PMID: 31669737. [DOI] [PubMed]
- 34.Foulcer S.J., Nelson C.M., Quintero M.V., Kuberan B., Larkin J., Dours-Zimmermann M.T., Zimmermann D.R., Apte S.S. Determinants of versican-V1 proteoglycan processing by the metalloproteinase ADAMTS5. J. Biol. Chem. 2014;289(40):27859–27873. doi: 10.1074/jbc.M114.573287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mjaatvedt C.H., Yamamura H., Capehart A.A., Turner D., Markwald R.R. The Cspg2 gene, disrupted in the hdf mutant, is required for right cardiac chamber and endocardial cushion formation. Dev. Biol. 1998;202(1):56–66. doi: 10.1006/dbio.1998.9001. PubMed PMID: 9758703. [DOI] [PubMed] [Google Scholar]
- 36.S. Nandadasa, A. O'Donnell, A. Murao, Y. Yamaguchi, R.J. Midura, L. Olson, S.S. Apte, The versican-hyaluronan complex provides an essential extracellular matrix niche for Flk1(+) hematoendothelial progenitors. Matrix Biol. 2021. Epub 2021/01/18. 10.1016/j.matbio.2021.01.002. PubMed PMID: 33454424. [DOI] [PMC free article] [PubMed]
- 37.Brown H.M., Dunning K.R., Robker R.L., Pritchard M., Russell D.L. Requirement for ADAMTS-1 in extracellular matrix remodeling during ovarian folliculogenesis and lymphangiogenesis. Dev. Biol. 2006;300(2):699–709. doi: 10.1016/j.ydbio.2006.10.012. PubMed PMID: 17097630. [DOI] [PubMed] [Google Scholar]
- 38.Russell D.L., Doyle K.M., Ochsner S.A., Sandy J.D., Richards J.S. Processing and localization of ADAMTS-1 and proteolytic cleavage of versican during cumulus matrix expansion and ovulation. J. Biol. Chem. 2003;278(43):42330–42339. doi: 10.1074/jbc.M300519200. PubMed PMID: 12907688. [DOI] [PubMed] [Google Scholar]
- 39.Shozu M., Minami N., Yokoyama H., Inoue M., Kurihara H., Matsushima K., Kuno K. ADAMTS-1 is involved in normal follicular development, ovulatory process and organization of the medullary vascular network in the ovary. J. Mol. Endocrinol. 2005;35(2):343–355. doi: 10.1677/jme.1.01735. PubMed PMID: 16216914. [DOI] [PubMed] [Google Scholar]
- 40.Dours-Zimmermann M.T., Maurer K., Rauch U., Stoffel W., Fassler R., Zimmermann D.R. Versican V2 assembles the extracellular matrix surrounding the nodes of ranvier in the CNS. J. Neurosci. 2009;29(24):7731–7742. doi: 10.1523/JNEUROSCI.4158-08.2009. PubMed PMID: 19535585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.I. Schechter, A. Berger, On the size of the active site in proteases. I. Papain. 1967. Biochem. Biophys. Res. Commun. 2012, 425(3):497-502. Epub 2012/08/29. 10.1016/j.bbrc.2012.08.015. PubMed PMID: 22925665. [DOI] [PubMed]
- 42.A.H. Beare, S. O'Kane, S.M. Krane, M.W. Ferguson, Severely impaired wound healing in the collagenase-resistant mouse. J Invest Dermatol. 2003, 120(1):153-63. Epub 2003/01/22. 12019 [pii] 10.1046/j.1523-1747.2003.12019.x. PubMed PMID: 12535212. [DOI] [PubMed]
- 43.Y. Fukumoto, J.O. Deguchi, P. Libby, E. Rabkin-Aikawa, Y. Sakata, M.T. Chin, C.C. Hill, P.R. Lawler, N. Varo, F.J. Schoen, S.M. Krane, M. Aikawa Genetically determined resistance to collagenase action augments interstitial collagen accumulation in atherosclerotic plaques. Circulation. 2004, 110(14):1953-9. Epub 2004/09/29. 10.1161/01.Cir.0000143174.41810.10. PubMed PMID: 15451791. [DOI] [PubMed]
- 44.R. Issa, X. Zhou, N. Trim, H. Millward-Sadler, S. Krane, C. Benyon, J. Iredale, Mutation in collagen-1 that confers resistance to the action of collagenase results in failure of recovery from CCl4-induced liver fibrosis, persistence of activated hepatic stellate cells, and diminished hepatocyte regeneration. FASEB J. 2003, 17(1):47-9. Epub 2002/12/12. 10.1096/fj.02-0494fje 02-0494fje [pii]. PubMed PMID: 12475903. [DOI] [PubMed]
- 45.X. Liu, H. Wu, M. Byrne, J. Jeffrey, S. Krane, R. Jaenisch, A targeted mutation at the known collagenase cleavage site in mouse type I collagen impairs tissue remodeling. J. Cell Biol. 1995, 130(1):227-37. Epub 1995/07/01. 10.1083/jcb.130.1.227. PubMed PMID: 7790374; PMCID: PMC2120510. [DOI] [PMC free article] [PubMed]
- 46.H. Wu, M.H. Byrne, A. Stacey, M.B. Goldring, J.R. Birkhead, R. Jaenisch, S.M. Krane, Generation of collagenase-resistant collagen by site-directed mutagenesis of murine pro alpha 1(I) collagen gene. Proc Natl Acad Sci U S A. 1990, 87(15):5888-92. Epub 1990/08/01. 10.1073/pnas.87.15.5888. PubMed PMID: 2165607; PMCID: PMC54434. [DOI] [PMC free article] [PubMed]
- 47.W. Zhao, M.H. Byrne, B.F. Boyce, S.M. Krane, Bone resorption induced by parathyroid hormone is strikingly diminished in collagenase-resistant mutant mice. J. Clin. Invest. 1999, 103(4):517-24. Epub 1999/02/18. 10.1172/jci5481. PubMed PMID: 10021460; PMCID: PMC408105. [DOI] [PMC free article] [PubMed]
- 48.W. Zhao, M.H. Byrne, Y. Wang, S.M. Krane, Osteocyte and osteoblast apoptosis and excessive bone deposition accompany failure of collagenase cleavage of collagen. J. Clin. Invest. 2000, 106(8):941-9. Epub 2000/10/18. 10.1172/jci10158. PubMed PMID: 11032854; PMCID: PMC314341. [DOI] [PMC free article] [PubMed]
- 49.Little C.B., Meeker C.T., Golub S.B., Lawlor K.E., Farmer P.J., Smith S.M., Fosang A.J. Blocking aggrecanase cleavage in the aggrecan interglobular domain abrogates cartilage erosion and promotes cartilage repair. J. Clin. Invest. 2007;117(6):1627–1636. doi: 10.1172/JCI30765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.E. Okugawa, H. Ogino, T. Shigenobu, Y. Yamakage, H. Tsuiji, H. Oishi, T. Kohno, M. Hattori, Physiological significance of proteolytic processing of Reelin revealed by cleavage-resistant Reelin knock-in mice. Sci. Rep. 2020, 10(1):4471. Epub 2020/03/13. 10.1038/s41598-020-61380-w. PubMed PMID: 32161359; PMCID: PMC7066138. [DOI] [PMC free article] [PubMed]
- 51.M. Egeblad, H.C. Shen, D.J. Behonick, L. Wilmes, A. Eichten, L.V. Korets, F. Kheradmand, Z. Werb, L.M. Coussens, Type I collagen is a genetic modifier of matrix metalloproteinase 2 in murine skeletal development. Dev Dyn. 2007, 236(6):1683-93. Epub 2007/04/19. 10.1002/dvdy.21159. PubMed PMID: 17440987; PMCID: PMC2581616. [DOI] [PMC free article] [PubMed]
- 52.Islam S., Chuensirikulchai K., Khummuang S., Keratibumrungpong T., Kongtawelert P., Kasinrerk W., Hatano S., Nagamachi A., Honda H., Watanabe H. Accumulation of versican facilitates wound healing: Implication of its initial ADAMTS-cleavage site. Matrix Biol. 2020;Epub 2019/11/02(87):77–93. doi: 10.1016/j.matbio.2019.10.006. PubMed PMID: 31669737. [DOI] [PubMed] [Google Scholar]
- 53.Carthy J.M., Meredith A.J., Boroomand S., Abraham T., Luo Z., Knight D., McManus B.M., Eickelberg O. Versican V1 Overexpression Induces a Myofibroblast-Like Phenotype in Cultured Fibroblasts. PLoS ONE. 2015;10(7):e0133056. doi: 10.1371/journal.pone.0133056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gueye N.A., Mead T.J., Koch C.D., Biscotti C.V., Falcone T., Apte S.S. Versican Proteolysis by ADAMTS Proteases and Its Influence on Sex Steroid Receptor Expression in Uterine Leiomyoma. J. Clin. Endocrinol. Metab. 2017;102(5):1631–1641. doi: 10.1210/jc.2016-3527. PubMed PMID: 28323982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mort J.S., Buttle D.J. The use of cleavage site specific antibodies to delineate protein processing and breakdown pathways. Mol Pathol. 1999;52(1):11–18. doi: 10.1136/mp.52.1.11. PubMed PMID: 10439833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.T.J. Mead, S.S. Apte Expression Analysis by RNAscope In Situ Hybridization. Methods Mol. Biol. 2020, 2043: 173-8. Epub 2019/08/30. Doi: 10.1007/978-1-4939-9698-8_14. PubMed PMID: 31463911. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.