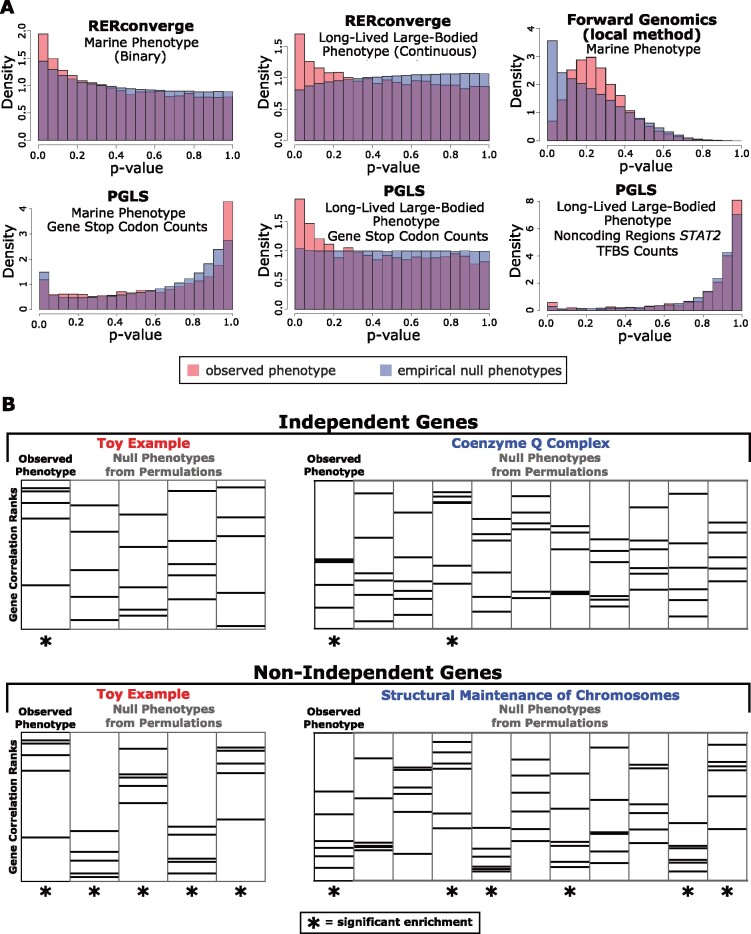
Fig. 1.
Permulations reveal statistical anomalies in genetic element- and pathway-level analyses because parametric P values deviate from the expected uniform distribution when assessed on null phenotypes. (A) P value histograms comparing P values obtained using an observed phenotype (red) compared with P values obtained from 500 (or more, see Results) null phenotypes from permulations. We evaluate a binary phenotype (marine) and a continuous phenotype (long-lived and large-bodied) through RERconverge, a binary phenotype (marine) through Forward Genomics, and a binary phenotype (marine) and a continuous phenotype (long-lived and large-bodied) through PGLS with gene stop codon counts and noncoding element STAT2 TFBS counts. In all cases, the empirical null from permulations (shown in blue) is nonuniform. Since null P value distributions are often nonuniform (shown in blue), observed parametric P values from standard statistical tests (shown in red) cannot be interpreted using traditional strategies. (B) Pathway enrichment statistics from RERconverge long-lived large-bodied analyses demonstrate artificially inflated significance because genes in many pathways are nonindependent. Accordingly, null phenotypes from permulations often show false signals of enrichment. Permulations correct for nonindependence by quantifying the frequency at which significant pathway enrichment occurs due to chance.