Abstract
Transcription factor-driven cell fate engineering in pluripotency induction, transdifferentiation, and forward reprogramming requires efficiency, speed, and maturity for widespread adoption and clinical translation. Here, we used Oct4, Sox2, Klf4, and c-Myc driven pluripotency reprogramming to evaluate methods for enhancing and tailoring cell fate transitions, through directed evolution with iterative screening of pooled mutant libraries and phenotypic selection. We identified an artificially evolved and enhanced POU factor (ePOU) that substantially outperforms wild-type Oct4 in terms of reprogramming speed and efficiency. In contrast to Oct4, not only can ePOU induce pluripotency with Sox2 alone, but it can also do so in the absence of Sox2 in a three-factor ePOU/Klf4/c-Myc cocktail. Biochemical assays combined with genome-wide analyses showed that ePOU possesses a new preference to dimerize on palindromic DNA elements. Yet, the moderate capacity of Oct4 to function as a pioneer factor, its preference to bind octamer DNA and its capability to dimerize with Sox2 and Sox17 proteins remain unchanged in ePOU. Compared with Oct4, ePOU is thermodynamically stabilized and persists longer in reprogramming cells. In consequence, ePOU: 1) differentially activates several genes hitherto not implicated in reprogramming, 2) reveals an unappreciated role of thyrotropin-releasing hormone signaling, and 3) binds a distinct class of retrotransposons. Collectively, these features enable ePOU to accelerate the establishment of the pluripotency network. This demonstrates that the phenotypic selection of novel factor variants from mammalian cells with desired properties is key to advancing cell fate conversions with artificially evolved biomolecules.
Keywords: reprogramming, protein engineering, POU, cell fate conversion, molecular evolution, transcription factor
Introduction
The forced overexpression of the transcription factor (TF) cocktail—Oct4, Sox2, Klf4, and c-Myc (OSKM) can reprogram somatic cells to pluripotency (Takahashi and Yamanaka 2006). Other TF combinations can directly interconvert somatic cell types in culture as well as in vivo (Heinrich et al. 2015). Cells generated by reprogramming methods enable disease modeling and drug testing, and could facilitate therapies through cell replacement. However, low success rates, lengthy procedures, safety and quality concerns as well as the failure to obtain mature cells are major bottlenecks impeding the clinical translation of cellular reprogramming technologies. Amongst the strategies to solve these problems are changes to the composition of TF cocktails (Di Stefano et al. 2014), supplementation of small molecules (Hou et al. 2013), modifications of culture conditions (Esteban et al. 2010), optimization of transgene delivery methods (Kim et al. 2016), and elimination of epigenetic roadblocks that impede the reprogramming process (Rais et al. 2013). We have proposed that tailoring the properties of the reprogramming TFs themselves through structure-based design is a powerful complementation to these efforts (Jauch et al. 2011; Jerabek et al. 2017). This idea was sparked by the finding that very subtle modifications to the DNA binding domains of TFs can dramatically change reprogramming outcomes. For example, a single missense mutation in Sox17 or Sox7 (generating factors termed Sox17EK and Sox7EK, respectively) can convert these genes that normally direct endodermal differentiation into potent inducers of pluripotent stem cells (iPSCs) (Jauch et al. 2011; Aksoy, Jauch, Eras, et al. 2013). Despite the initial success to improve cellular reprogramming through rational protein design, its scope was limited because of an incomplete understanding of the structural basis for chromatin recognition and the protein interaction networks in the nucleus. We thus asked whether directed molecular evolution can help in the identification of improved reprogramming factors. Directed evolution strategies are common in biotechnology and enzyme enhancement (Arnold 2018). Yet, it is not normally used in the engineering of gene regulatory networks in mammalian cells. In an initial study, we could show that pooled screens with saturation mutagenesis libraries enabled the isolation of pluripotency reprogramming factors based on the scaffolds of Sox2 and Sox17 that outperform their wild-type (WT) counterparts (Veerapandian et al. 2018).
Here, we applied our protein engineering and directed evolution paradigm to the Pit-Oct-Unc (POU) TF family. We evaluated an iterative screening strategy that combined saturation mutagenesis and domain shuffling. We used the POU member Oct4, as it is a key factor in pluripotency reprogramming and in the maintenance of embryonic stem cells (ESC). Of the four Yamanaka factors, Sox2, Klf4, and c-Myc could be replaced by paralogous genes, but Oct4 could not be substituted by other POU family members such as Oct1 or Oct6 when using retroviral reprogramming in mouse (Nakagawa et al. 2008; Jerabek et al. 2017; Malik et al. 2019).
We show that directed evolution in mammalian cells using pooled screens with randomized TF libraries, cell selection, and sequencing led to the identification of an artificially evolved and enhanced POU (ePOU) that outperformed Oct4 by an order of magnitude in the mouse reprogramming system. We found that subtle modifications to the DNA binding landscape of POU factors and their biophysical stabilization contribute to the orchestration of regulatory networks, which in effect leads to a more forceful penetration of epigenetic barriers. Given the manifold global influence of these seemingly subtle modifications on protein function, we conclude that readouts based on cellular phenotypes are the key to isolate enhanced reprogramming factors. We anticipate that analogous experimental evolution strategies will help to eliminate bottlenecks that impede transdifferentiation and forward programming systems in vitro and in vivo.
Results
Generation of an Artificially Evolved POU Factor That Outperforms Oct4 in Pluripotency Reprogramming
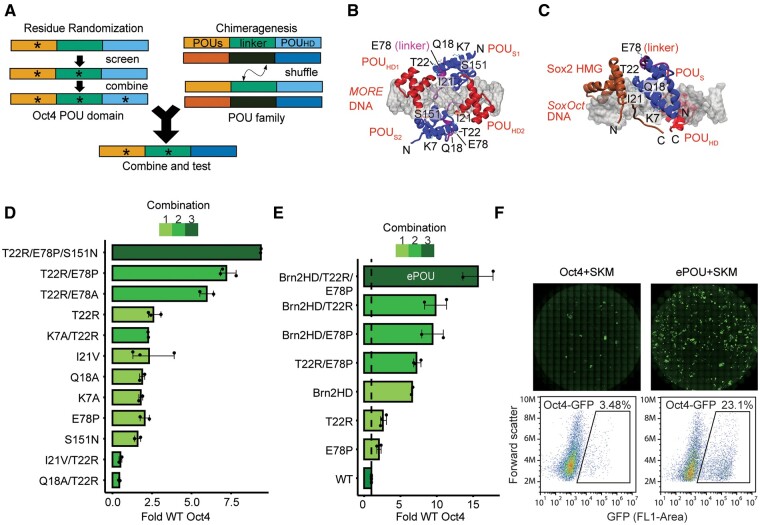
To evaluate whether the scaffold of the POU family of TFs can be improved through directed molecular evolution in mammalian cells, we chose the generation of iPSCs from mouse embryonic fibroblasts (MEFs) (Takahashi and Yamanaka 2006) as a benchmark. Under standard serum/LIF conditions, <0.1% of starting MEF cells can be successfully reprogrammed to iPSCs using the retroviral overexpression of Oct4, Sox2, Klf4, and c-Myc (OSKM) transgenes. In mice, Oct4 cannot be replaced by paralogous POU family factors, such as Oct6 and Brn2. We decided to employ site saturation mutagenesis at selected amino acid positions in Oct4 and the recombination of functional protein domains (“chimeragenesis”) in our search for functionally improved POU factors (fig. 1A, supplementary fig. S1A–C, Supplementary Material online). Oct4 libraries were constructed by randomizing six amino acids selected by analyzing protein interaction interfaces in structural models (fig. 1B and C), evolutionary conservation patterns (supplementary fig. S1D, Supplementary Material online), and functional effects in previous studies (Nishimoto et al. 2005; Esch et al. 2013; Jerabek et al. 2017). We performed screens in MEFs carrying a GFP transgene driven by the Oct4 promoter (OG2-MEFs) that allows the detection of emergent pluripotent cells through the activation of the GFP reporter (Szabo et al. 2002). We aspired to sample the total sequence space of the six amino acid positions, but the large size of a combinatorial library (206 = 64 million sequence variants) precluded a combinatorial screen. We thus opted to screen six individual point mutation libraries in parallel and replaced WT Oct4 to induce pluripotency in combination with Sox2 (S), Klf4 (K), and c-Myc (M) (supplementary fig. S1A, Supplementary Material online). Mutant transgenes producing iPSC colonies were extracted by PCR, genotyped by sequencing, and scored for their ability to outperform Oct4 (supplementary fig. S1E, Supplementary Material online). We next combined top candidates in an iterative fashion and observed profound synergies for subsets of variant combinations (supplementary fig. S1B and fig. 1D, Supplementary Material online). Chimeragenesis libraries were generated by recombining components of the tripartite DNA binding POU domain (POU specific domain, POUS, linker and POU homeodomain, POUHD) between Oct4 and the somatic POU factors Brn2, Oct6, and Oct1 by way of single-tube USER® enzyme recombination (supplementary fig. S1C, Supplementary Material online). The top performers from site saturation mutagenesis and chimeragenesis libraries outperformed WT Oct4 by 2-fold and 7-fold, respectively (fig. 1D, supplementary fig. S1E and F, Supplementary Material online). A combination of a T22R/E78P double mutant with a chimeric protein containing the POUHD of Brn2 within the scaffold of Oct4 was identified as the most potent reprogramming factor from our screen (fig. 1E). We termed this factor “ePOU” for artificially evolved and enhanced POU TF. ePOU has a total of 17 amino acid substitutions compared with WT Oct4 and generated approximately 15 times more iPSC colonies than WT Oct4 (fig. 1E and F). ePOU-derived iPSCs lines no longer expressed transgenes and did not encode transgenic Oct4 (supplementary fig. S2A–C, Supplementary Material online). iPSC lines expressed Sox2, Oct4 and Nanog (supplementary fig. S2D, Supplementary Material online), and showed a normal karyotype (supplementary fig. S2E, Supplementary Material online). ePOU and Oct4 derived iPSCs could be cultured in Serum/LIF, chemically defined naïve (2i/LIF) and primed (FGF2/ActivinA) pluripotency culture conditions with indistinguishable morphology (supplementary fig. S2F–G, Supplementary Material online). We next verified pluripotency and differentiation potential of ePOU derived iPSC lines both in vivo and in vitro. Early passage ePOU iPSCs injected into mouse blastocysts were able to generate chimeric mice that grew into adulthood (supplementary fig. S3A, Supplementary Material online). In vitro differentiation experiments showed that ePOU iPSCs could differentiate into the three germ layers reminiscent to Oct4 iPSCs (supplementary fig. S3B and C and supplementary movies S5, S6, Supplementary Material online). In sum, we conclude ePOU-derived iPSCs are bona fide pluripotent cells. Collectively, the versatile scaffold of the bipartite POU domain is amenable to functional enhancements in pluripotency reprogramming by protein design.
Fig. 1.
Identification of an evolved and enhanced POU factor through iterative phenotypic selection. (A) Scheme of the iterative screen to evolve the Oct4 scaffold by residue randomization and POU family chimeragenesis. (B, C) Structural model of (B) the Oct4 homodimer on MORE DNA and (C) a Sox2/Oct4 heterodimer on the SoxOct DNA. The six mutated residues (K7, Q18, I21, T22, E78, and S151) are labeled when visible. (D) iPSC colony count data relative to WT Oct4 for selected single, double, and triple combinations of mutations. (E) iPSC colony count data relative to WT Oct4 for chimeric POU factors point mutated variants and combinations thereof. The variant composed of the Brn2-POUHD plus T22R/E78P performed best. The variant was subsequently termed “ePOU” and selected for further characterization. (F) Whole-well scans showing Oct4-GFP+ colonies (upper panel) and cells sorted for GFP fluorescence (lower panel) using FACS. In (D–E), data are shown as mean of 2–3 biological replicates (three technical replicates each) with the range shown as error bars. POUS, POU Specific; POUHD, POU Homeodomain; HMG, High Mobility Group; O, Oct4; S, Sox2; K, Klf4; M, c-Myc; MORE, More palindromic Oct factor Recognition Element; iPSC, induced pluripotent stem cells.
ePOU Rescues Reprogramming Conditions Where Oct4 Is Inactive
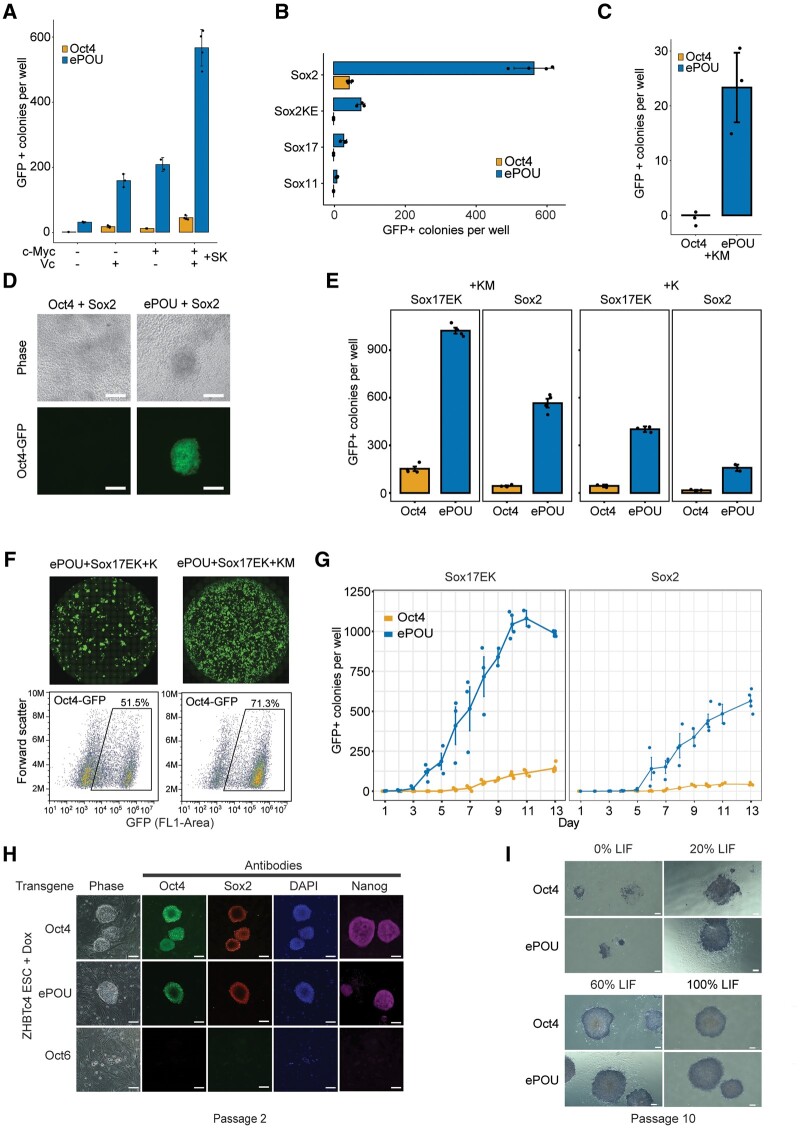
We went on to explore whether ePOU can support iPSC generation under conditions in which Oct4 performs poorly or is unable to reprogram. Omitting c-Myc and vitamin C led to an overall reduction in the yield of iPSC colonies. Yet under these conditions, ePOU still outperformed Oct4 in a pronounced manner (fig. 2A). Oct4 requires Sox2 to induce pluripotency. When Sox2 is replaced with Sox11, Sox17, or Sox2KE (a Sox2 mutant defective in Oct4 heterodimerization), Oct4 containing cocktails cannot generate iPSCs (Nakagawa et al. 2008; Jauch et al. 2011). However, ePOU effectively rescued reprogramming conditions with these Sox factors (fig. 2B). In fact, Sox factors could be omitted altogether when Oct4 is replaced by ePOU (fig. 2C). Moreover, two-factor ePOU/Sox2 cocktails could generate iPSC colonies in serum/LIF with vitamin C whereas the Oct4/Sox2 pair failed to do so (fig. 2D). This demonstrates that ePOU supports iPSC generation under conditions in which Oct4 cannot.
Fig. 2.
ePOU outperforms Oct4 in a range of reprogramming conditions. (A–C) Colony count data of pluripotency reprogramming experiments: (A) in the absence or presence of c-Myc and Vitamin C, (B) using Sox factors that compromise iPSC generation in the presence of WT Oct4 (Sox2KE, Sox17, and Sox11) in four-factor conditions, and (C) Three-factor conditions omitting Sox factors. (D) Oct4-GFP+ fluorescence images of iPSC colonies generated with two-factor reprogramming of Oct4+Sox2 (left) and ePOU+Sox2 (right) cocktails (scale = 100µm). (E) Oct4-GFP+ colony count data for reprogramming experiments to evaluate synergies of eSOX (Sox17EK) and ePOU in the absence or presence of c-Myc. (F) Whole well Oct4-GFP fluorescence scans and fluorescence-activated cell sorting (FACS) with GFP channel for ePOU/Sox17EK combinations in the absence or presence of c-Myc. (G) Time course of GFP+ colony counts from day 1 to day 13 in four-factor conditions. (H) Colonies of ZHBTc4 ESCs transduced with Oct4, ePOU, or Oct6 in the presence of Dox and 100% LIF (10 ng/ml) after two passages, stained with indicated antibodies (scale = 80 µm). (I) Alkaline phosphatase staining of ZHBTc4 ESC colonies transduced with Oct4 or ePOU at varying concentrations of LIF at passage 10 (scale = 200µm). In (A–C, E), data are mean ± SEM of three biological replicates with 2–3 technical replicates each. Colonies are counted at day 13 of reprogramming. O, Oct4; S, Sox2; K, Klf4; M, c-Myc; Sox2KE, Sox2 K57E mutant defective in Oct4 heterodimerization; Sox17EK, Sox17 E57K mutant with enhanced pluripotency reprogramming capacity; ESC, embryonic stem cells; LIF, Leukemia inhibitory factor; Dox, doxycycline.
Collaboration between Re-Engineered Sox and POU Factors
We next examined whether ePOU-driven reprogramming could be further boosted with engineered Sox factors. We have previously shown that Sox17EK, a point-mutated Sox17 variant with a single glutamate-to-lysine substitution within its DNA binding domain, turns into a potent inducer of pluripotency (Jauch et al. 2011; Ng et al. 2012; Aksoy, Jauch, Chen, et al. 2013). Here, we found that the use of ePOU/Sox17EK combinations led to a further increase of the colony yield compared with ePOU/Sox2 (fig. 2E and F). Four-factor ePOU/Sox17EK/K/M cocktails resulted in an efficiency of up to 15% (iPSC colonies/plated MEFs; supplementary fig. S3D, Supplementary Material online). More than 70% of viable cells were GFP+ by day 13 (fig. 2F). ePOU/Sox17EK cocktails profoundly accelerated iPSC generation, with the first GFP+ cells appearing on day 3, with those in WT Oct4/Sox2 controls typically appearing on days 6–7 post-transduction (fig. 2G, supplementary movies S1–4, Supplementary Material online). We conclude that artificial factors can collaborate with one another. Thus, components of reprogramming factors’ cocktails could be optimized concurrently for tailored cell fate engineering strategies.
ePOU Retains the Capacity to Maintain Pluripotency
After activation of the pluripotency network, exogenous transgenes, such as ePOU, are naturally silenced in iPSCs and no longer impact cellular properties. We wondered whether ePOU would modify the function of Oct4 during the self-renewal of ESCs. We thus performed a pluripotency maintenance assay using the ZHBTc4 ESC line (Niwa et al. 2000). In these cells, endogenous Oct4 is substituted by a tet-off Oct4 transgene that can be repressed with doxycycline (Dox) leading to loss of self-renewal and differentiation. When cultured under serum/LIF conditions in the presence of Dox, pluripotency was rescued by ePOU and Oct4 but not by Oct6 (fig. 2H). When we compared Oct4 and ePOU in serum/LIF conditions, we observed similar cellular morphologies, marker expression, and alkaline phosphatase (AP) staining patterns even with gradual LIF withdrawal (fig. 2I, supplementary fig. S3E, Supplementary Material online). In the absence of LIF, exogenous Oct4 is insufficient to support prolonged maintenance of ESCs (He et al. 2017). Correspondingly, extended passaging in the absence of LIF led to a loss of pluripotency for both ePOU and Oct4 conditions (supplementary fig. S3F, Supplementary Material online). Together, we conclude that the various changes in ePOU compared with Oct4 did not debilitate its capacity to support self-renewal.
ePOU Speeds Up Reprogramming without Changing the Route to Pluripotency
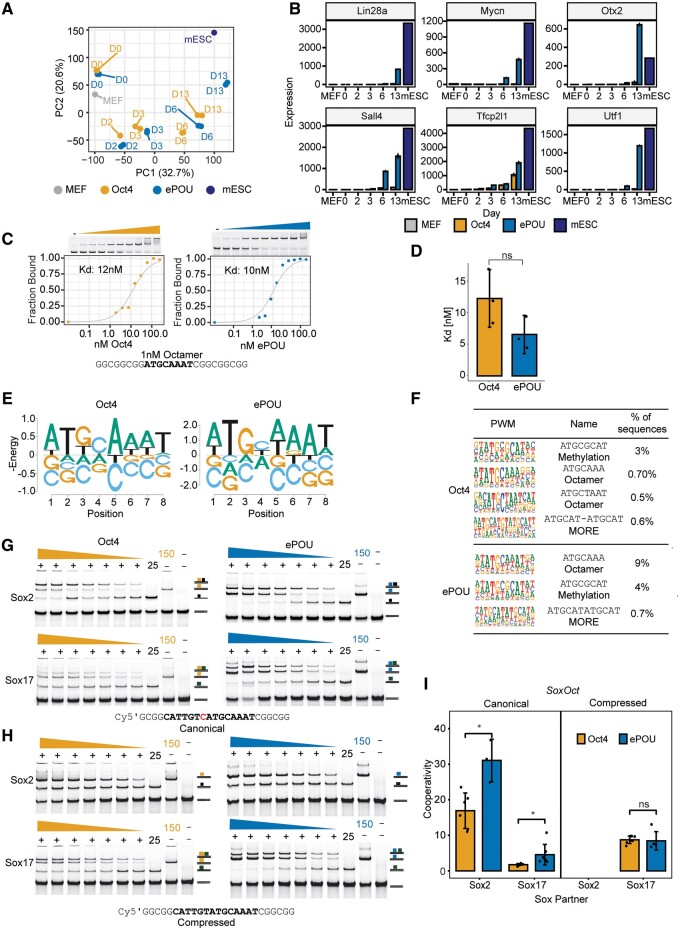
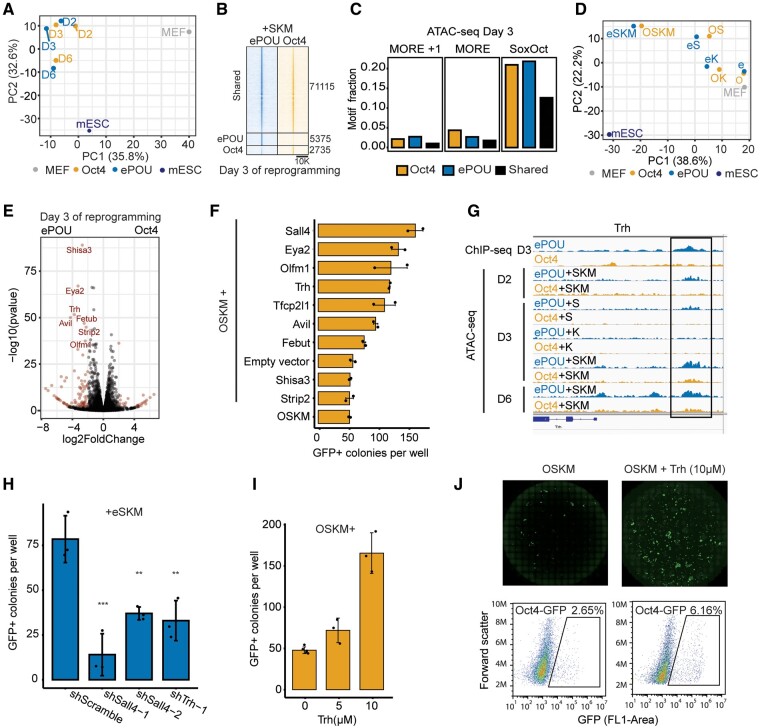
Several pluripotency reprogramming systems were found to take different paths (Polo et al. 2012; Velychko, Adachi, et al. 2019) and passed through alternative transitory states before converging at the common iPSC endpoint (Cheng et al. 2019). We performed time-course RNA sequencing to determine whether Oct4 and ePOU-expressing cells are subject to different reprogramming routes. We analyzed RNA samples collected from reprogramming cells at days 0, 2, 3, 6, and 13 expressing ePOU or Oct4 in four-factor conditions. We observed that the overall trajectory of global gene expression profiles was similar, but compared with Oct4, the bulk of ePOU expressing cells approximate the ESC state at earlier time points (fig. 3A). Consistently, we found that a subset of differentially expressed genes upregulated earlier and more strongly in ePOU-expressing cells are involved in embryonic pathways (supplementary fig. S4A and B, Supplementary Material online). Notable pluripotency related genes upregulated include Lin28a, Mycn, Oxt2, Sall4, Tfcp2l1, and Utf1 (fig. 3B). Together, ePOU activates the pluripotency network more quickly without major divergences from the standard route to pluripotency.
Fig. 3.
ePOU accelerates reprogramming without switching reprogramming trajectories, monomeric DNA specificity or Sox partners. (A) Principal-component analysis (PCA) of global gene expression profiles determined by RNA-seq for cells transduced with Oct4 or ePOU+SKM at days 0, 2, 3, 6, and 13 along with public data sets of MEFs (GSE103979) (Malik et al. 2019) and mESC (GSE93029) (Li et al. 2017). (B) Expression of six selected pluripotency-related genes. (C, D) (C) Binding of ePOU and Oct4 to canonical octamer DNA. Varying protein concentrations (0–500 nM) were incubated with 1 nM fluorescently labeled DNA. Binding isotherms and dissociation constants (Kd) are shown under the gels. (D) Kd’s from three independent titration EMSAs. (E) Energy logos derived from Spec-seq using a set of sequences with one nucleotide difference to the canonical octamer motif (ATGCAAAT). Note that the vertical axis is -Energy so that the highest affinity bases are on top and each column is normalized to a mean of 0. (F) Top enriched motifs in the third cycle of high throughput-SELEX for Oct4 and ePOU. Motifs shown are the octamer, MORE and methylation motif (a palindromic motif with a CpG methylation site). (G–I) Heterodimer EMSAs with 50 nM Cy5 labeled DNA probes for (G) canonical and (H) compressed SoxOct DNA elements to monitor the complex formation of ePOU (blue) or Oct4 (orange) with Sox2 (black square) and Sox17 (green square). (I) Quantifications of heterodimer EMSAs and calculation of cooperativity factors according to (Ng et al. 2012). In (D, I), data are shown as mean ± SD (n = 3–5). * P-value < 0.05 from an unpaired t-test. MEF, mouse embryonic fibroblasts; mESC, mouse embryonic stem cells; MORE, More palindromic Oct factor Recognition Element.
Specificity for Octamer DNA and Cobinding with Sox Partners Is Similar for ePOU and Oct4
To test if differences in DNA binding between ePOU and Oct4 contribute to enhanced reprogramming, we purified recombinant proteins of their respective DNA-binding domains (supplementary fig. S4C, Supplementary Material online). Both proteins bound to the canonical octamer DNA with similar high affinity (fig. 3C and D). To rigorously analyze the specificity for DNA sequences, we established two complementary high-throughput assays. We first performed Specificity by sequencing (Spec-seq) for the quantitative profiling of the relative binding affinities of 100’s–1000’s octamer-like sequences in parallel (Stormo et al. 2015). Second, we performed high-throughput systematic evolution of ligands by exponential enrichment (HT-SELEX) using a 40 N random library to comprehensively probe the sequence space to identify the most preferred sequences but without direct quantitative information (Jolma et al. 2010). Spec-seq was done using three libraries where four nucleotides of the octamer DNA were randomized (supplementary fig. S4D, Supplementary Material online). Relative binding energies to the sampled sequence space correlated strongly for ePOU and Oct4 (supplementary fig. S4E, Supplementary Material online), which led to similar energy logos for the top-scoring sequences (fig. 3E). ePOU had a wider range of energies in comparison to Oct4, suggesting that it might be more specific for the consensus compared with Oct4 (supplementary fig. S4E, Supplementary Material online). HT-SELEX was performed using His6-tagged proteins in four rounds of enrichment. We recovered a similar set of top-scoring motifs from the sequence sets enriched by ePOU or Oct4, respectively, albeit with flipped ranks (fig. 3F). Motifs include versions of the canonical octamer as well as two palindromic motifs: first, the MORE (More palindromic Oct factor Recognition Element) motif that we and others had previously found to contribute to the cistrome of Oct4 (Mistri et al. 2015; Jerabek et al. 2017) and second, a compact motif that was reported to be effectively bound by Oct4, preferably so when the central CpG dinucleotide is methylated (Yin et al. 2017).
Oct4 forms DNA-dependent heterodimers with Sox2 in pluripotent cells and with Sox17 in the extraembryonic endoderm (Jauch et al. 2011; Aksoy, Jauch, Chen, et al. 2013). The dimerization in pluripotent cells is driven by a composite SoxOct element termed the canonical motif where Sox and Oct half-sites are juxtaposed in a defined manner. The Sox17/Oct4 dimerization in extraembryonic endoderm occurs on a variant “compressed motif” where a single nucleotide at the 3′ end of the Sox half-site is removed. The altered arrangement of the Sox and Oct half-sites switches the protein interfaces that facilitate the cooperative partnership between Sox and POU factors. To analyze whether ePOU interacts differently with Sox2 and Sox17, we performed Electrophoretic mobility shift assays (EMSAs) using either the canonical or compressed SoxOct DNA elements. We observed similar dimerization patterns with Sox factors for both proteins (fig. 3G, H). For the compressed element, ePOU and Oct4 could not form heterodimers with Sox2, indicating competitive binding. However, both POU factors showed positive cooperativity with Sox17 (fig. 3H and I). Conversely, on the canonical element, Oct4 and ePOU exhibited strongly positive cooperativity with Sox2. They could also form heterodimers with Sox17 but with significantly reduced efficiency (fig. 3G and I). For both Sox factors, we recorded moderately elevated cooperativity constants for ePOU compared with Oct4 with the canonical motif (fig. 3I). In sum, core functions of the POU domain, such as the preference for the octamer element and dimerization patterns with Sox factors are largely preserved between ePOU and Oct4.
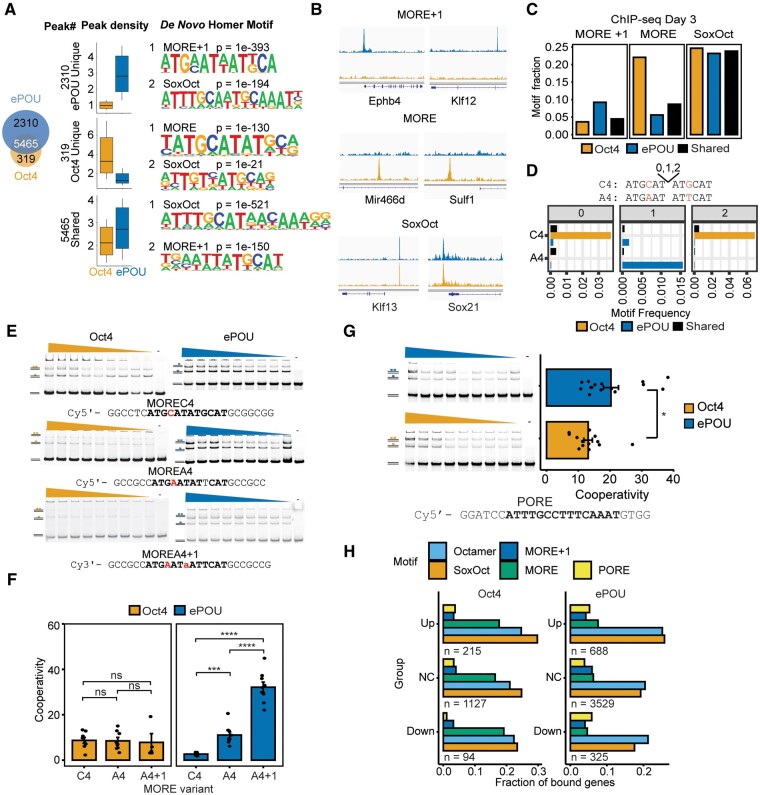
ePOU Preferentially Binds and Homodimerizes on a Palindromic MORE+1 Motif
To compare chromatin association of ePOU and Oct4 at an early phase of reprogramming, we performed chromatin immunoprecipitation sequencing (ChIP-seq) at day 3. We found that ePOU not only bound the majority of Oct4 targeted loci, but also bound a sizable number of unique sites (fig. 4A). By contrast, Oct4 was only bound to a small set of unique sites. This suggests that the enhanced reprogramming efficiencies by ePOU might be mediated by the effects that ePOU has on these additional sites (fig. 4B). De novo motif discovery and motif scanning showed that the canonical SoxOct motif was the top hit in the shared sites and was also strongly enriched in unique sites which is consistent with the strong cooperativity with Sox2 for both factors in EMSA (fig. 4A). Interestingly, there was a differential enrichment of variants of the palindromic MORE motifs in the uniquely bound peak sets (fig. 4C and D). MORE motifs facilitate homodimeric binding of POU factors with varying degrees of cooperativity (Rhee et al. 1998; Jerabek et al. 2017) and are enriched at binding sites of somatic POU factors, such as Brn2 and Oct6 (Malik et al. 2018). MORE motifs can be classified by the spacing between the palindromic half-sites (0, 1, or 2 base pair spacers) and the identity of the base at position four of the core motif (C4 vs. A4) (Mistri et al. 2015). The MORE + 1 motif with an A4 was enriched in peaks uniquely bound by ePOU. Oct4, however, preferentially bound the spacer-less C4 version of the MORE. (fig. 4D). To test whether these differences are intrinsic properties of the respective POU domains, we determined homodimer cooperativity factors using EMSAs (fig. 4E and F). We observed indistinguishable moderate cooperativity for Oct4 on the three tested MORE variants. However, ePOU bound MORE A4 + 1 with significantly higher cooperativity than MORE C4 (fig. 4F). Interestingly, a subset of ePOU-specific MORE+1 binding sites are located at endogenous retroviruses of the RLTR13 subfamily (supplementary fig. S5A and B, Supplementary Material online). The RLTR13 family plays roles in gene regulation of ESCs and trophoblast stem cells (Todd et al. 2019). We verified the homodimerization of ePOU to this element (supplementary fig. S5C, Supplementary Material online).
Fig. 4.
ePOU preferentially targets a MORE motif variant as a homodimer. (A) Left: Venn diagram for ChIP-seq peaks of Oct4 and ePOU at day 3 of reprogramming defined using MAnorm (Shao et al. 2012). Center: Normalized Oct4 and ePOU ChIP-seq signals. Right: Top two de novo motifs for each peak category. Boxes represent the interquartile range with a horizontal median line. Whiskers extend up to 1.5 times the interquartile range. (B) Genome browser tracks of Oct4 and ePOU ChIP-seq peaks at selected gene loci containing matches to indicated motifs. Genomic coordinates are listed in supplementary table S5, Supplementary Material online. (C, D) Motif occurrences within each peak category in A determined by (C) PWM scanning and (D) text search with perfect matches to MORE subtypes A4 and C4 with spacers 0–2. (E–G) (E) EMSAs using MORE variants MOREC4, MOREA4, and MOREA4 + 1. Dimeric and monomeric states are marked. (F) Homodimer cooperativity factors determined by densitometric analysis as in (Jerabek et al. 2017). (G) EMSAs using PORE DNA and corresponding homodimer cooperativity factors. Data are shown as mean ± SEM (n = 5–15). *, ***, **** P-value <0.05, 0.001, and 0.0001, respectively from an unpaired t-test with Benjamini–Hochberg correction. EMSAs were performed with 50 nM DNA and protein concentrations of 0–400 nM. (H) Fractional presence of DNA motifs within Oct4 (left) and ePOU (right) ChIP-seq peaks nearby genes that are upregulated (Up), not changed (NC) or downregulated (Down) at reprogramming day 6 with respect to MEFs. MORE, More Oct factor Recognition Element.
To obtain insight into the molecular basis for the specific DNA preferences of ePOU, we constructed structural models of ternary ePOU/MORE A4 + 1 and Oct4/MORE C4 complexes (supplementary fig. S5D, Supplementary Material online). None of the 17 modified amino acids are in the immediate vicinity of the protein-DNA contact interface, yet the modifications profoundly change the surface charge for substitutions within the POUHD. It was previously noted that the majority of divergent mutations within the POU family occur at POUHD helices some of which are proximal to the homodimerization interface and might modulate binding to palindromic DNA (Gold et al. 2014).
The differential binding to MORE elements inspired us to search for other previously reported homodimerization motifs, such as PORE (Palindromic Octamer Recognition Element) (Botquin et al. 1998) and NORE (N-Oct-3 Responsive Element; N-Oct-3 is a synonym for Brn2) (Alazard et al. 2005; Nieto et al. 2007) in our ChIP-seq peaks. Motif scanning showed that PORE and NORE were slightly more enriched at sites uniquely bound by ePOU compared with Oct4 sites (supplementary fig. S5E, Supplementary Material online). Likewise, ePOU has a moderately higher cooperativity than Oct4 on PORE in EMSAs (fig. 4G). On NORE, only ePOU was able to form homodimers whereas Oct4 binds as monomer (supplementary fig. S5F, Supplementary Material online).
We next asked whether the binding of ePOU and Oct4 in the context of alternative motifs would lead to a differential association with upregulated, downregulated and unchanged (NC) genes at day 6 of reprogramming. We found that the octamer and SoxOct motifs are most prevalently associated with ePOU and Oct4 bound genes in particular if they are upregulated (fig. 4H). PORE/MORE + 1 motifs are often linked to ePOU bound genes whereas MORE binding occurs in a higher fraction of Oct4 bound genes regardless of the transcriptional response of the nearby genes.
Collectively, we conclude that sequence substitutions within the POU domain bias the preference of ePOU toward the MORE + 1, PORE, and NORE elements. This newly acquired property did not compromise the capacity of ePOU to bind functionally critical octamer and SoxOct elements. We surmise that the acquired preference for a distinct set of composite DNA elements (such as those linked to RLTR13 repeats) contributes to the enhancement of ePOU-driven cell fate conversions.
ePOU Retains a Limited Capacity to Open Chromatin
A key feature of reprogramming TFs is their ability to access and open silenced chromatin. Sox2, Oct4, and Klf4 can bind nucleosome core particles (NCPs) in vitro (Soufi et al. 2015). The binding mechanism of Sox2 and Oct4 has been structurally characterized for NCPs encompassing synthetic DNA sequences (Michael et al. 2020). To test whether enhanced reprogramming by ePOU is mediated by optimized closing of somatic genes and/or opening of pluripotency genes, we performed ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing) at different reprogramming stages and after transducing different factor combinations. Overall, the dynamics of chromatin changes were similar in all tested ePOU and Oct4 conditions when four factors were used (fig. 5A). Less than 10% of the loci were differentially opened on day 3, with the vast majority of sites opening concurrently (fig. 5B). As in ChIP-seq, shared opening loci were strongly enriched for SoxOct motifs whereas MORE and MORE + 1 motifs are more prevalent in Oct4 or ePOU sites, respectively (fig. 5C, supplementary fig. S6A, Supplementary Material online).
Fig. 5.
ePOU differentially activates novel reprogramming facilitators but does not gain pioneering activity. (A) PCA of ATAC-seq signals at indicated reprogramming stages for four-factor experiments. (B) ATAC-seq signals around shared and unique ATAC-seq peaks defined using MAnorm (Shao et al. 2012) at day 3 in four-four factor experiments. (C) Motif occurrences within each class of ATAC-seq peaks defined in (B). (D) PCA of ATAC-seq signals for cells transduced with ePOU or Oct4 alone, as 2F cocktail with Sox2, 2F cocktail with Klf4 or as part of four-factor cocktails at day 3 of reprogramming. (E) Volcano plot highlighting differentially expressed genes in four-factor ePOU versus Oct4 conditions at day 3. Dots are red when P < 0.05 and |log2FoldChange| > 2. (F) Colony count data for reprogramming experiments with OSKM along with candidates selected from (E). Data shown is mean ± range (n = 2). (G) Genome browser tracks of Trh loci showing ChIP-seq and ATAC-seq signals. Location of a MORE+1 motif is indicated with a black bar. Coordinates are in supplementary table S5, Supplementary Material online. (H) Effect of shRNA knock-down on the Oct4-GFP+ colony number at day 13 in ePOU-SKM conditions. Cells were cultured in the presence of 1 µg/ml puromycin to select for cells expressing indicated shRNAs. Data shown as mean ± SD (n = 3) **, *** P ≤ 0.01, 0.001 from Tukey’s test after ANOVA. (I, J) (I) Oct4-GFP+ colonies obtained using OSKM in the presence of 0, 5, or 10 μM TRH peptide in the medium. Data shown as mean ± SD (n = 3–5). (J) The corresponding whole-well scans and cell sorting results. O, Oct4; e, ePOU; S, Sox2; K, Klf4; M, c-Myc; MEF, mouse embryonic fibroblasts; mESC, mouse embryonic stem cells; D2, day 2; D3, day3; D6, day6.
When ePOU and Oct4 are expressed alone without Sox2 or Klf4, the chromatin state at day 3 of reprogramming was barely altered in comparison to MEFs (fig. 5D). Yet, the addition of Klf4 or Sox2 triggers profound chromatin changes that were further amplified when the four factors were jointly expressed (fig. 5D, supplementary fig. S6B and C, Supplementary Material online). These observations are consistent with our recent findings that Sox2 by itself drives chromatin opening more potently than Oct4 (Malik et al. 2019). Likewise, when inspecting the subset of sites annotated as MEF enhancers (Shen et al. 2012), we found that Oct4 and ePOU alone induced minimal changes but given the presence of Sox2 or Klf4, ePOU accelerated the closing of these somatic sites (supplementary fig. S6B, Supplementary Material online). Sox2 and Oct4 can cooperate in the context of NCPs (Michael et al. 2020) and Oct4 amplifies Sox2-driven chromatin opening (Malik et al. 2019). ePOU appears to preserve this function of Oct4 but is not markedly enhanced in its role as a pioneering factor that facilitates chromatin opening.
ePOU Selectively Activates Novel Facilitators of Reprogramming
Global RNA-seq analysis suggested overall similar reprogramming routes for Oct4- and ePOU-expressing cells. To identify individual genes that are activated in an ePOU-specific manner, we performed differential gene expression analysis (fig. 5E, supplementary fig. S6D, Supplementary Material online). The genes activated earlier and more potently in ePOU included known pluripotency factors, such as Sall4 and Tfcp2l1 (fig. 3B). In addition, there were genes previously not implicated to play a role in pluripotency (Shisa3, Avil, Eya2, Fetub, Olfm1, Strip2, and Trh) (fig. 5E, supplementary fig. S6D, Supplementary Material online). To test whether these genes act as reprogramming facilitators, we expressed them alongside OSKM to generate iPSCs. We found that five of seven novel factors could significantly boost reprogramming efficiency (fig. 5F). We became interested in Trh as it contained proximal binding sites with ePOU specific ChIP-seq and ATAC-seq signals (fig. 5G). Trh encodes for the precursor of a secreted tripeptide called thyrotropin-releasing hormone (TRH), which acts as a neurotransmitter in the hypothalamus, contributes to metabolic regulation as well as neuromodulation of cardiac and respiratory function (Frohlich and Wahl 2019). It is expressed in embryonic and epiblast stem cells (McKnight et al. 2007) and was found to be moderately upregulated during pluripotency reprogramming in mice (Bansho et al. 2017). To further validate the effect of Trh, we performed shRNA knockdown of Trh alongside Sall4 as a positive control and observed a 2–3-fold reduction in iPSC colony numbers (fig. 5H, supplementary fig. S6E and F, Supplementary Material online). We next supplemented reprogramming media with the processed TRH peptide and found that reprogramming efficiency increases ∼3-fold (fig. 5I and J). Collectively, these findings demonstrate that the improved potency of ePOU relies on the differential regulation of a novel set of reprogramming facilitators including the neurotransmitter TRH.
ePOU Is Thermally Stabilized and Resists Cellular Degradation
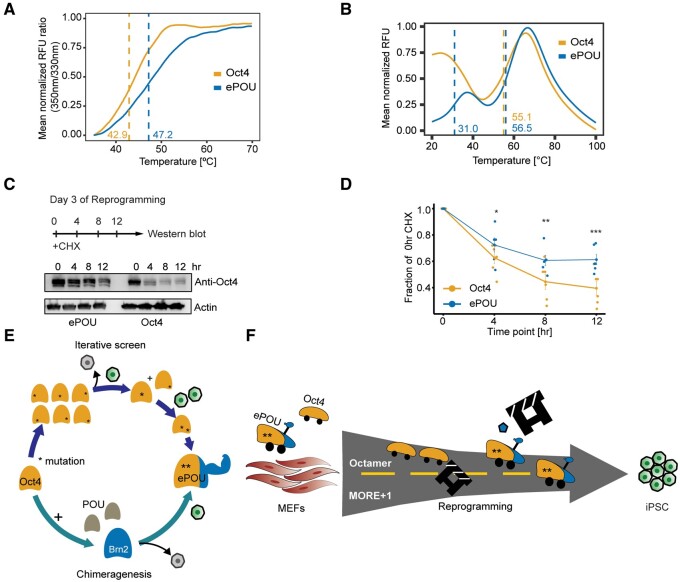
Destabilizing mutations and mutations that reduce cellular abundance are a major cause for the malfunction of disease-associated proteins (Yue et al. 2005; Matreyek et al. 2018). Conversely, the enhancement of thermal stability has led to the successful directed evolution of enzymes and antibodies with superior performance compared with their wild type counterparts (Asial et al. 2013). Mutants that affect post-translational modification (PTM) sites (phosphorylation or ubiquitination) of Oct4 increasing its stability have been shown to improve iPSC generation (Bae et al. 2017; Li et al. 2018). Alternatively, loss-of-function Oct4 mutations show decreased stability (Jin et al. 2016). We therefore decided to analyze the unfolding kinetics of the purified POU domains of Oct4 and ePOU using two complementary differential scanning fluorometry (DSF) assays. First, we performed nanoscale DSF (nanoDSF). NanoDSF is a dye-free technique that relies on intrinsic tryptophan fluorescence to detect the unfolding of proteins. ePOU showed a stronger resistance to thermal unfolding with an increase in melting temperature by 4 °C (fig. 6A). The second DSF assay is based on the interaction of hydrophobic patches that become exposed during unfolding with the Sypro®Orange dye. Oct4 showed a high fluorescent signal already at the starting temperature (20 °C), followed by a second unfolding transition with a melting temperature (Tm) of 55 °C (fig. 6B). ePOU mimicked the second unfolding transition with a slightly increased Tm of 56.5 °C. In stark contrast to Oct4, ePOU underwent an initial unfolding transition ∼30 °C. We reason that the bimodal unfolding is due to the bipartite structure of the POU domain and thus envisage two possible trajectories. On one hand, POUHD and POUS could unfold sequentially in an independent manner. On the other hand, POUHD and POUS may interact intramolecularly. The resolution of this interaction as well as the subsequent unfolding of the subdomains were both gauged in the experiment. Either way, the high initial signal indicates that Oct4 is profoundly floppier compared with the structurally stabilized ePOU showing a distinct two-step unfolding kinetics.
Fig. 6.
ePOU is stabilized in comparison to Oct4. (A B) Normalized thermal unfolding curves of purified ePOU and Oct4 DNA binding domains determined using (A) nanoDSF based on intrinsic tryptophan fluorescence and (B) fluorescence emission of a Sypro® Orange dye. Unfolding transitions were measured thrice and melting points (Tm) indicated by dashed lines were estimated using the peak of the first derivative of the melt curve. (C, D) Cycloheximide (CHX) chase assay in reprogramming MEFs transfected with Oct4 or ePOU +SKM at day 3. (C) Representative western blot using an Oct4 antibody with actin as control. (D) Quantification of Oct4/ePOU immunoblot bands (normalized for Actin). *, **, *** P ≤ 0.05, 0.01, and 0.001, respectively from an unpaired t-test. Data are shown as mean ± SEM (n = 6, 2 biological replicates). (E) Schema for the iterative screen with phenotypic selection leading to the discovery of the ePOU. Oct4 (orange), Brn2 (blue), ePOU (orange+blue with an arm to symbolize increased stability), and gray (other POU factors). Green cells are GFP+ iPSCs whereas gray cells are nonreprogrammed ones. * indicates point mutations. (F) ePOU has two-point mutations (asterisk) as well as a fragment from Brn2. ePOU accelerates reprogramming compared with Oct4 by acquiring new binding preferences (i.e., MORE+1) while retaining binding sites of Oct4. Increased robustness/enhanced stability allows ePOU more effective removal of the roadblock and accompanied by expression of factors, such as TRH (pentagon). MEFs, mouse embryonic fibroblasts; iPSC, induced pluripotent stem cells.
We next probed how the biophysical stabilization affects protein abundance in reprogramming cells. To this end, we performed a cycloheximide chase assay on reprogramming day 3, a time point at which endogenous Oct4 protein is still repressed (fig. 6C). We observed that Oct4 protein levels declined significantly faster than ePOU levels (fig. 6D). Thus, ePOU is thermodynamically stabilized compared with Oct4, leading to a longer-lived protein with reduced turnover. This may in turn prolong residence times at enhancers, reinforce transcriptional cascades and consequently boost its capacity to reprogram.
Discussion
To engineer cell fates, genetic methods based on exogenously introduced factor cocktails remain the favored choice to induce pluripotency, to directly transdifferentiate somatic cells, and to forward program stem and progenitor cells toward mature cells. Typically, genes that play prominent roles in the desired target cell types are chosen under the assumption that such factors are best suited to guide the starting cells toward a path leading to successful cell fate conversions. Although this assumption has led to seminal breakthroughs in the stem cell field, the overall procedure of cell fate conversion is amenable for improvements by a multitude of ways. Importantly, many reprogramming procedures lead to nonnatural cell fate switches that do not happen in vivo. Therefore, reprogramming TFs have not been subject to natural selection pressure to orchestrate this artificial process. Consistently, reprogramming methods have been established based on factors that lack a function in the target cell type (Montserrat et al. 2013; Shu and Deng 2013). We have shown that the somatic genes Sox17 and Oct6 can become pluripotency-inducing factors with a few point mutations. In the case of engineered Sox17, a high-performance factor was produced that substantially outperforms Sox2 (Jauch et al. 2011). However, in the case of re-engineered Oct6, it fails to reach the levels of its paralogue Oct4 (Jerabek et al. 2017). Here, we provide a proof of concept that directed factor evolution can dramatically improve biomolecule driven cell fate conversions beyond Sox. We report on ePOU to demonstrate that the scaffold of POU factors can be artificially evolved through an iterative screen involving saturation mutagenesis and chimeragenesis (fig. 6E). ePOU not only outperforms Oct4 by an order of magnitude (even more so when combined with Sox17EK), but it also speeds up reprogramming and enables pluripotency induction when otherwise essential components are omitted.
The identification and characterization of ePOU presented here hold several lessons for protein and stem cell engineering, as well as the mechanism of biomolecule driven reprogramming (fig. 6F). First, if biochemical methods are considered for TF engineering, they should not focus on optimizing affinity to cognate binding elements, such as the octamer. The global binding profile spanning low, medium, and high-affinity sites contribute to the genome-wide regulatory profile necessary for phenotypic transitions. The accommodation of target genes linked to the MORE + 1 element is likely important and yet, could not have been predicted.
Second, protein stability and prolonged half-life are relevant parameters amenable for optimization. We do not posit that this leads to a mere increase in nuclear protein levels. Rather, it may affect turnover, dynamics within (super) enhancers, and the reinforcement of transcriptional output. Protein stability is also related to PTM. ePOU modifications do not directly alter any PTM sites but are situated directly beside these sites (supplementary fig. S7, Supplementary Material online). These mutations may indirectly affect recognition sites of modifying enzymes. For example, the putative kinase recognition site for S229 is K R T S229 I E N R where N232/R233 are mutated in ePOU. The redesign of PTM sites could further improve reprogramming factors.
Third, the effects of point mutations on factor activity are hard to predict and are nonadditive. Results from single-site mutagenesis screens only improved reprogramming outcomes by a maximum of 1.5–2-fold. Yet, the effects of having combinatorial nonadditive effects proved to be profound. Likewise, previous domain swap experiments suggested that the POUHD domain had minimal effects on reprogramming (Velychko, Kang, et al. 2019). Yet, our redefinition of domain boundaries changed the outcome, with the POUHD fragment of the neural Brn2 factor significantly contributing to the enhanced factor.
Lastly, directed evolution experiments should be carried out in cellular system and species intended to be improved. ePOU accelerates pluripotency reprogramming in mouse but is unable to outperform human OCT4 (data not shown). This is likely because newly acquired features, such as the specific binding to mouse specific retrotransposons of the RLTR13 family are not expected to help in the regulation of human genes. Similar evolutionary differences between other POU reprogramming factors have been observed (Kim et al. 2020). OCT6 is unable to support retroviral reprogramming in mice. In human, however, OCT6 could reproducibly generate iPSCs. This emphasizes that the cis-regulatory genomic landscape differs between species. Directed evolution experiments could lead to a species-specific adaptation of the evolving factor to drive cell state conversions.
We conclude that contextually relevant phenotypic readouts are superior to in vitro evaluation (i.e., the affinity to cognate binding sites by phage display or related methods) when evolving factors. Complementary to phenotypic readouts, biophysical assays that monitor protein stability (Asial et al. 2013) or the propensity to form molecular condensates (Sabari et al. 2020), could be considered. Ultimately, phenotypic readouts that stringently assess factor variants and intelligent variant libraries are most promising to generate otherwise refractory cell types. It is clearly desirable to minimize screening variability by ensuring single integration events at safe harbor loci. Still, the stochastic nature of the reprogramming process creates challenges to assign a meaningful function score to each variant in the factor library. Assays that probe protein abundance or cell viability are less stochastic than the overall complex phenomenon of cellular reprogramming (Matreyek et al. 2018). Nevertheless, it is highly desirable to comprehensively probe every amino acid of a given TF, perhaps using readouts, such as single cell gene expression profiles (Datlinger et al. 2017). Lessons learned from deep mutational scans (Matreyek et al. 2018) would complement directed evolution of reprogramming factors by cell selection and sequencing (DERBY-seq) (Veerapandian et al. 2018) which uses combinatorial libraries and cannot comprehensibly probe >3–5 positions per screen.
Collectively, by applying principles of directed evolution in mammalian cells, we demonstrate that native factors can be profoundly enhanced to accelerate reprogramming and to successfully convert cells where wild type factors fail. This strategy can straightforwardly be applied to lineage reprogramming in vitro or in vivo as well as for the forward programming of stem and progenitor cells. We surmise that the success of methods based on artificially evolved and enhanced transcription factors (eTFs) is only limited by the availability of suitable phenotypic selection strategies to ensure arrival at the desired destination.
Materials and Methods
Cell Culture
MEFs were obtained from embryonic day 13.5 embryos of OG2 mice carrying a GFP transgene driven by the Oct4 promoter (Szabo et al. 2002). The OG2-MEFs were cultured in MEF medium. Plat-E packaging cells (Morita et al. 2000) were maintained in DMEM/10% FBS. Reprogramming experiments were performed in mouse ESC medium. Pluripotent cells were maintained using Serum/LIF or cultured in 2i/LIF medium (Ying et al. 2008). Cells were cultured at 37 °C and 5% CO2. Further details and media compositions are in the supplementary extended methods.
Retrovirus Production and Reprogramming
Retroviral infection and reprogramming were done as described in (Veerapandian et al. 2018; Malik et al. 2019; Srivastava et al. 2020). Briefly, before transduction, Plat-E cells were seeded at 8 × 106 cells per 10 cm plate. On the next day, 10 μg retroviral pMX vector and 40 μg linear polyethyleneimine dissolved in 1 ml Opti-MEM were added. The medium was replaced after 12–14 h and virus-containing supernatants were collected at 48 h and 72 h, filtered, diluted to 12 ml with Plat-E medium, and mixed with polybrene (Sigma–Aldrich; #40804ES76). MEFs were seeded and transduced with 0.5 ml of each freshly prepared viral supernatant. The second round of infections was done 24 h later and after 24 h the viral supernatant was replaced with mouse ESC medium containing 50 μg/ml Vitamin C unless indicated otherwise. This day was defined as reprogramming day 0 and the medium was replaced daily on subsequent days. GFP+ colonies were counted from the moment of appearance until day 13. Fluorescence-activated cell sorting (FACS) was performed to quantify GFP+ cells after trypsinization using 10,000 life cells. To establish iPSC lines, cells from iPSC colonies were picked at day 13 into 1.5 ml tubes and transferred into 24-well plates precoated with feeders. The cells were grown for ∼5 days until sizeable iPSC colonies developed and picking was repeated. After two rounds of colony picking, cells were passaged by trypsination. New reprogramming factors were synthesized by Guangzhou IGE biotechnology and cloned into the pMXs-Flag vector (supplementary table S3, Supplementary Material online). 0.5 ml retroviral supernatant was used together with OSKM and 0.5 ml pMXs-Flag or Plat-E cell medium were controls. EPOU/S/K/M iPSC colonies were picked up on day 7 and used to generate chimeric mice. To measure efficiency of iPSCs induction at different cellular densities, OG2-MEF cells were diluted by 50% percent 7 times in different wells of 12-well dishes, and iPSC colonies were counted at day 13.
Protein Expression and Purification
Expression vector pET28a-ePOU was constructed through amplifying the POU domain of the pMX-ePOU and was ligated into pET28a using XhoI and NdeI sites (supplementary table S1, Supplementary Material online). Constructs with an N-terminal His6x tag were expressed in Escherichia coli Rosetta (DE3) competent cells grown in LB medium + 0.2% glucose and 100 µg/ml kanamycin at 37 °C to OD600 of ∼0.6–0.8 before adding 0.5 mM isopropyl-beta-thio galactosidase (IPTG) for induction at 16–18 °C for 18–22 h. The cell pellet was resuspended in lysis buffer and incubated for 30 min on ice before freezing at –80 °C. Cells were thawed and disrupted by sonification. Sonication was done for 10 min at 50% amplitude for 8 s on/8s off and centrifuged His tagged fusion proteins were first captured using HisPur Ni-NTA Superflow Agarose and eluted with elution buffer. The elute was exchanged for desalting buffer via the PD10 desalting column. Proteins were further purified using an ÄKTA pure and a 1 or 5 ml HiTrap Heparin column equilibrated in desalting buffer and eluted with a linear NaCl gradient to 1 M NaCl. Samples are then further purified with HiLoad 16/600 Superdex 75 column with the storage buffer. Proteins were concentrated using the centrifugal filter units and stored at –80 °C. The high mobility group (HMG) domains of Sox2 and Sox17 were expressed and purified as described in (Ng et al. 2012; Srivastava et al. 2020). Buffer details are included in the extended methods.
Electrophoretic Mobility Shift Assay
Double‐stranded DNA (dsDNA) probes with 5′ Cy5 or Cy3 dyes attached to the forward strand were prepared using an annealing buffer (20 mM Tris/HCl, 50 mM MgCl2, 50 mM KCl, pH 8.0). Protein samples and fluorescently labeled DNA are incubated for ∼2 h in EMSA buffer (10 mM Tris/HCl pH 8.0, 0.1 mg ml − 1 BSA, 50 µM ZnCl2, 100 mM KCl, 10% glycerol, 0.10% Igepal CA630, 2 mM βME). Gels were first preran using a 1 × TG buffer (Tris 0.25 mM, glycine 192 mM, pH 8.0) at 200 V for 30 min. Then, 10 µl samples were loaded and gels were run for 30 min at 200 V at 4 °C. Images are captured using an Amersham Typhoon 5 Biomolecular Imager and quantified using ImageQuantTL 7.0. DNA probes used are listed in supplementary table S4, Supplementary Material online. Cooperativity factors were calculated as described in (Ng et al. 2012) for heterodimers and (Jerabek et al. 2017; Wang et al. 2018) for homodimers.
Extended Methods
Detailed experimental procedures for cell culture, mutagenesis, genotyping, karyotyping, chimeric mice generation, cyclohexamide chase assay, Spec-seq, HT-SELEX, ChIP-seq, ATAC-seq, RNA-seq, pluripotency maintenance, knockdowns, spontaneous differentiation, DSF, and bioinformatics analysis are available in the supplementary extended methods.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgements
R.J. is supported by the National Natural Science Foundation of China (Grant No. 31771454), the Research Grants Council of Hong Kong General Research Fund (RGC/GRF) projects number 17128918 and 17101120, a Health and Medical Research Fund (06174006), and the Germany/Hong Kong Joint Research Scheme sponsored by the Research Grants Council of Hong Kong and the German Academic Exchange Service (Reference No. G-HKU701/18). We thank Shih Chieh Jeff Ti for his help in purifying proteins, Pik Fan Wong for admistrative support, and Leon Li for access to Nanotemper Tycho NT.6.
Author Contributions
D.S.T. and Y.C. contributed to study design, performed most experiments, and analyzed data. Y.C. performed the iterative screens leading to the identification of ePOU. V.M. performed ChIP-seq experiments. X.Y., Y.G., A.B., S.V., Y.W., and M.W. contributed to reprogramming experiments. A.B. and D.S.T. performed and analyzed stability assays. Y.G. and A.B. performed knock-down experiments, Y.G., S.Y.H., and A.B performed pluripotency maintenance assays. Y.G. and D.H.H.H. performed in vitro differentiation assay. Y.S. performed structural modeling. Y.W. performed ATAC-seq experiments. D.S.T., Y.W., and V.M. performed bioinformatics analysis. H.R.S. supervised reprogramming experiments of S.V. and analyzed data. F.L. and D.S.T. performed HT-SELEX experiments supervised by J.Y., G.D.S. contributed to the design and analysis of Spec-seq experiments. R.J. designed and supervised the study, acquired funding, and analyzed data. D.S.T., Y.C., and R.J. wrote the manuscript with input from all authors.
Data Availability
The data underlying this article are available in Gene Expression Omnibus at https://www.ncbi.nlm.nih.gov/geo/ under the accession numbers GSE104219 (RNA-seq), GSE104220 (ChIP-seq), and GSE161512 (ATAC-seq).
References
- Aksoy I, Jauch R, Chen J, Dyla M, Divakar U, Bogu GK, Teo R, Leng Ng CK, Herath W, Lili S, et al. 2013. Oct4 switches partnering from Sox2 to Sox17 to reinterpret the enhancer code and specify endoderm. EMBO J. 32(7):938–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksoy I, Jauch R, Eras V, Chng WB, Chen J, Divakar U, Ng CK, Kolatkar PR, Stanton LW.. 2013. Sox transcription factors require selective interactions with oct4 and specific transactivation functions to mediate reprogramming. Stem Cells 31(12):2632–2646. [DOI] [PubMed] [Google Scholar]
- Alazard R, Blaud M, Elbaz S, Vossen C, Icre G, Joseph G, Nieto L, Erard M.. 2005. Identification of the ‘NORE’ (N-Oct-3 responsive element), a novel structural motif and composite element. Nucleic Acids Res. 33(5):1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold FH. 2018. Directed evolution: bringing new chemistry to life. Angew Chem Int Ed Engl. 57(16):4143–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asial I, Cheng YX, Engman H, Dollhopf M, Wu B, Nordlund P, Cornvik T.. 2013. Engineering protein thermostability using a generic activity-independent biophysical screen inside the cell. Nat Commun. 4:2901. [DOI] [PubMed] [Google Scholar]
- Bae KB, Yu DH, Lee KY, Yao K, Ryu J, Lim DY, Zykova TA, Kim MO, Bode AM, Dong Z.. 2017. Serine 347 phosphorylation by JNKs negatively regulates OCT4 protein stability in mouse embryonic stem cells. Stem Cell Rep. 9(6):2050–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansho Y, Lee J, Nishida E, Nakajima-Koyama M.. 2017. Identification and characterization of secreted factors that are upregulated during somatic cell reprogramming. FEBS Lett. 591(11):1584–1600. [DOI] [PubMed] [Google Scholar]
- Botquin V, Hess H, Fuhrmann G, Anastassiadis C, Gross MK, Vriend G, Scholer HR.. 1998. New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes Dev. 12(13):2073–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Pei Y, He L, Peng G, Reinius B, Tam PPL, Jing N, Deng Q.. 2019. Single-cell RNA-seq reveals cellular heterogeneity of pluripotency transition and X chromosome dynamics during early mouse development. Cell Rep. 26(10):2593–2607.e3. [DOI] [PubMed] [Google Scholar]
- Datlinger P, Rendeiro AF, Schmidl C, Krausgruber T, Traxler P, Klughammer J, Schuster LC, Kuchler A, Alpar D, Bock C.. 2017. Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods. 14(3):297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano B, Sardina JL, van Oevelen C, Collombet S, Kallin EM, Vicent GP, Lu J, Thieffry D, Beato M, Graf T.. 2014. C/EBPalpha poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature 506(7487):235–239. [DOI] [PubMed] [Google Scholar]
- Esch D, Vahokoski J, Groves MR, Pogenberg V, Cojocaru V, Vom Bruch H, Han D, Drexler HC, Arauzo-Bravo MJ, Ng CK, et al. 2013. A unique Oct4 interface is crucial for reprogramming to pluripotency. Nat Cell Biol. 15(3):295–301. [DOI] [PubMed] [Google Scholar]
- Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J, Li W, Weng Z, Chen J, Ni S, et al. 2010. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 6(1):71–79. [DOI] [PubMed] [Google Scholar]
- Frohlich E, Wahl R.. 2019. The forgotten effects of thyrotropin-releasing hormone: metabolic functions and medical applications. Front Neuroendocrinol. 52:29–43. [DOI] [PubMed] [Google Scholar]
- Gold DA, Gates RD, Jacobs DK.. 2014. The early expansion and evolutionary dynamics of POU class genes. Mol Biol Evol. 31(12):3136–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R, Xhabija B, Al-Qanber B, Kidder BL.. 2017. OCT4 supports extended LIF-independent self-renewal and maintenance of transcriptional and epigenetic networks in embryonic stem cells. Sci Rep. 7(1):16360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C, Spagnoli FM, Berninger B.. 2015. In vivo reprogramming for tissue repair. Nat Cell Biol. 17(3):204–211. [DOI] [PubMed] [Google Scholar]
- Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, et al. 2013. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science 341(6146):651–654. [DOI] [PubMed] [Google Scholar]
- Jauch R, Aksoy I, Hutchins AP, Ng CK, Tian XF, Chen J, Palasingam P, Robson P, Stanton LW, Kolatkar PR.. 2011. Conversion of Sox17 into a pluripotency reprogramming factor by reengineering its association with Oct4 on DNA. Stem Cells 29(6):940–951. [DOI] [PubMed] [Google Scholar]
- Jerabek S, Ng CK, Wu G, Arauzo-Bravo MJ, Kim KP, Esch D, Malik V, Chen Y, Velychko S, MacCarthy CM, et al. 2017. Changing POU dimerization preferences converts Oct6 into a pluripotency inducer. EMBO Rep. 18(2):319–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Wang L, Zhu F, Tan W, Lin W, Chen D, Sun Q, Xia Z.. 2016. Critical POU domain residues confer Oct4 uniqueness in somatic cell reprogramming. Sci Rep. 6:20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Kivioja T, Toivonen J, Cheng L, Wei G, Enge M, Taipale M, Vaquerizas JM, Yan J, Sillanpaa MJ, et al. 2010. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 20(6):861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KP, Wu Y, Yoon J, Adachi K, Wu G, Velychko S, MacCarthy CM, Shin B, Ropke A, Arauzo-Bravo MJ, et al. 2020. Reprogramming competence of OCT factors is determined by transactivation domains. Sci Adv. 6(36):eaaz7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SI, Oceguera-Yanez F, Sakurai C, Nakagawa M, Yamanaka S, Woltjen K.. 2016. Inducible transgene expression in human iPS cells using versatile all-in-one piggyBac transposons. Methods Mol Biol. 1357:111–131. [DOI] [PubMed] [Google Scholar]
- Li D, Liu J, Yang X, Zhou C, Guo J, Wu C, Qin Y, Guo L, He J, Yu S, et al. 2017. Chromatin accessibility dynamics during iPSC reprogramming. Cell Stem Cell. 21(6):819–833.e6. [DOI] [PubMed] [Google Scholar]
- Li S, Xiao F, Zhang J, Sun X, Wang H, Zeng Y, Hu J, Tang F, Gu J, Zhao Y, et al. 2018. Disruption of OCT4 ubiquitination increases OCT4 protein stability and ASH2L-B-mediated H3K4 methylation promoting pluripotency acquisition. Stem Cell Rep. 11(4):973–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik V, Glaser LV, Zimmer D, Velychko S, Weng M, Holzner M, Arend M, Chen Y, Srivastava Y, Veerapandian V, et al. 2019. Pluripotency reprogramming by competent and incompetent POU factors uncovers temporal dependency for Oct4 and Sox2. Nat Commun. 10(1):3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik V, Zimmer D, Jauch R.. 2018. Diversity among POU transcription factors in chromatin recognition and cell fate reprogramming. Cell Mol Life Sci. 75(9):1587–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matreyek KA, Starita LM, Stephany JJ, Martin B, Chiasson MA, Gray VE, Kircher M, Khechaduri A, Dines JN, Hause RJ, et al. 2018. Multiplex assessment of protein variant abundance by massively parallel sequencing. Nat Genet. 50(6):874–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight KD, Hou J, Hoodless PA.. 2007. Dynamic expression of thyrotropin-releasing hormone in the mouse definitive endoderm. Dev Dyn. 236(10):2909–2917. [DOI] [PubMed] [Google Scholar]
- Michael AK, Grand RS, Isbel L, Cavadini S, Kozicka Z, Kempf G, Bunker RD, Schenk AD, Graff-Meyer A, Pathare GR, et al. 2020. Mechanisms of OCT4-SOX2 motif readout on nucleosomes. Science 368(6498):1460–1465. [DOI] [PubMed] [Google Scholar]
- Mistri TK, Devasia AG, Chu LT, Ng WP, Halbritter F, Colby D, Martynoga B, Tomlinson SR, Chambers I, Robson P, et al. 2015. Selective influence of Sox2 on POU transcription factor binding in embryonic and neural stem cells. EMBO Rep. 16(9):1177–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montserrat N, Nivet E, Sancho-Martinez I, Hishida T, Kumar S, Miquel L, Cortina C, Hishida Y, Xia Y, Esteban CR, et al. 2013. Reprogramming of human fibroblasts to pluripotency with lineage specifiers. Cell Stem Cell. 13(3):341–350. [DOI] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T.. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7(12):1063–1066. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S.. 2008. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 26(1):101–106. [DOI] [PubMed] [Google Scholar]
- Ng CK, Li NX, Chee S, Prabhakar S, Kolatkar PR, Jauch R.. 2012. Deciphering the Sox-Oct partner code by quantitative cooperativity measurements. Nucleic Acids Res. 40(11):4933–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto L, Joseph G, Stella A, Henri P, Burlet-Schiltz O, Monsarrat B, Clottes E, Erard M.. 2007. Differential effects of phosphorylation on DNA binding properties of N Oct-3 are dictated by protein/DNA complex structures. J Mol Biol. 370(4):687–700. [DOI] [PubMed] [Google Scholar]
- Nishimoto M, Miyagi S, Yamagishi T, Sakaguchi T, Niwa H, Muramatsu M, Okuda A.. 2005. Oct-3/4 maintains the proliferative embryonic stem cell state via specific binding to a variant octamer sequence in the regulatory region of the UTF1 locus. Mol Cell Biol. 25(12):5084–5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG.. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 24(4):372–376. [DOI] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. 2012. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151(7):1617–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, et al. 2013. Deterministic direct reprogramming of somatic cells to pluripotency. Nature 502(7469):65–70. [DOI] [PubMed] [Google Scholar]
- Rhee JM, Gruber CA, Brodie TB, Trieu M, Turner EE.. 1998. Highly cooperative homodimerization is a conserved property of neural POU proteins. J Biol Chem. 273(51):34196–34205. [DOI] [PubMed] [Google Scholar]
- Sabari BR, Dall'Agnese A, Young RA.. 2020. Biomolecular condensates in the nucleus. Trends Biochem Sci. 45(11):961–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Zhang Y, Yuan GC, Orkin SH, Waxman DJ.. 2012. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol. 13(3):R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488(7409):116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu J, Deng H.. 2013. Lineage specifiers: new players in the induction of pluripotency. Genomics Proteomics Bioinformatics 11(5):259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS.. 2015. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161(3):555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava Y, Tan DS, Malik V, Weng M, Javed A, Cojocaru V, Wu G, Veerapandian V, Cheung LWT, Jauch R.. 2020. Cancer-associated missense mutations enhance the pluripotency reprogramming activity of OCT4 and SOX17. FEBS J. 287(1):122–144. [DOI] [PubMed] [Google Scholar]
- Stormo GD, Zuo Z, Chang YK.. 2015. Spec-seq: determining protein-DNA-binding specificity by sequencing. Brief Funct Genomics. 14(1):30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo PE, Hubner K, Scholer H, Mann JR.. 2002. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech Dev. 115(1–2):157–160. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S.. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126(4):663–676. [DOI] [PubMed] [Google Scholar]
- Todd CD, Deniz O, Taylor D, Branco MR.. 2019. Functional evaluation of transposable elements as enhancers in mouse embryonic and trophoblast stem cells. Elife 8:e44344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerapandian V, Ackermann JO, Srivastava Y, Malik V, Weng M, Yang X, Jauch R.. 2018. Directed evolution of reprogramming factors by cell selection and sequencing. Stem Cell Rep. 11(2):593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velychko S, Adachi K, Kim KP, Hou Y, MacCarthy CM, Wu G, Scholer HR.. 2019. Excluding Oct4 from Yamanaka cocktail unleashes the developmental potential of iPSCs. Cell Stem Cell. 25(6):737–753.e734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velychko S, Kang K, Kim SM, Kwak TH, Kim KP, Park C, Hong K, Chung C, Hyun JK, MacCarthy CM, et al. 2019. Fusion of reprogramming factors alters the trajectory of somatic lineage conversion. Cell Rep. 27(1):30–39.e4. [DOI] [PubMed] [Google Scholar]
- Wang X, Srivastava Y, Jankowski A, Malik V, Wei Y, Del Rosario RC, Cojocaru V, Prabhakar S, Jauch R.. 2018. DNA-mediated dimerization on a compact sequence signature controls enhancer engagement and regulation by FOXA1. Nucleic Acids Res. 46(11):5470–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, et al. 2017. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356(6337):eaaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A.. 2008. The ground state of embryonic stem cell self-renewal. Nature 453(7194):519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P, Li Z, Moult J.. 2005. Loss of protein structure stability as a major causative factor in monogenic disease. J Mol Biol. 353(2):459–473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in Gene Expression Omnibus at https://www.ncbi.nlm.nih.gov/geo/ under the accession numbers GSE104219 (RNA-seq), GSE104220 (ChIP-seq), and GSE161512 (ATAC-seq).