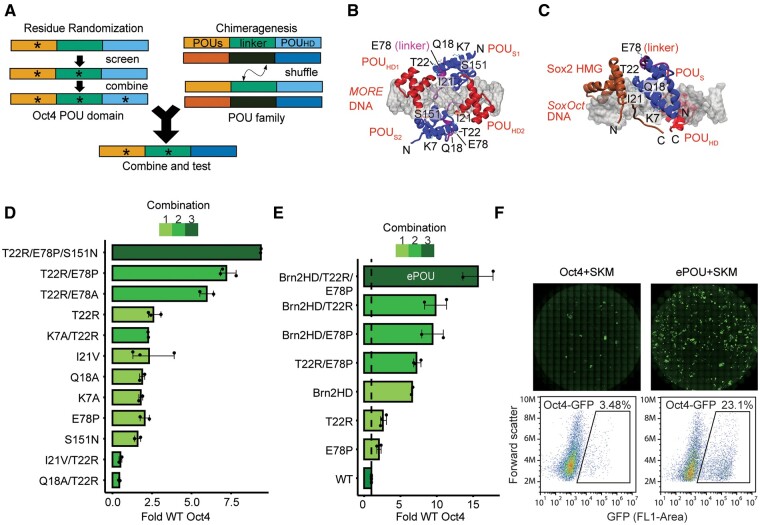
Fig. 1.
Identification of an evolved and enhanced POU factor through iterative phenotypic selection. (A) Scheme of the iterative screen to evolve the Oct4 scaffold by residue randomization and POU family chimeragenesis. (B, C) Structural model of (B) the Oct4 homodimer on MORE DNA and (C) a Sox2/Oct4 heterodimer on the SoxOct DNA. The six mutated residues (K7, Q18, I21, T22, E78, and S151) are labeled when visible. (D) iPSC colony count data relative to WT Oct4 for selected single, double, and triple combinations of mutations. (E) iPSC colony count data relative to WT Oct4 for chimeric POU factors point mutated variants and combinations thereof. The variant composed of the Brn2-POUHD plus T22R/E78P performed best. The variant was subsequently termed “ePOU” and selected for further characterization. (F) Whole-well scans showing Oct4-GFP+ colonies (upper panel) and cells sorted for GFP fluorescence (lower panel) using FACS. In (D–E), data are shown as mean of 2–3 biological replicates (three technical replicates each) with the range shown as error bars. POUS, POU Specific; POUHD, POU Homeodomain; HMG, High Mobility Group; O, Oct4; S, Sox2; K, Klf4; M, c-Myc; MORE, More palindromic Oct factor Recognition Element; iPSC, induced pluripotent stem cells.