Figure 1.
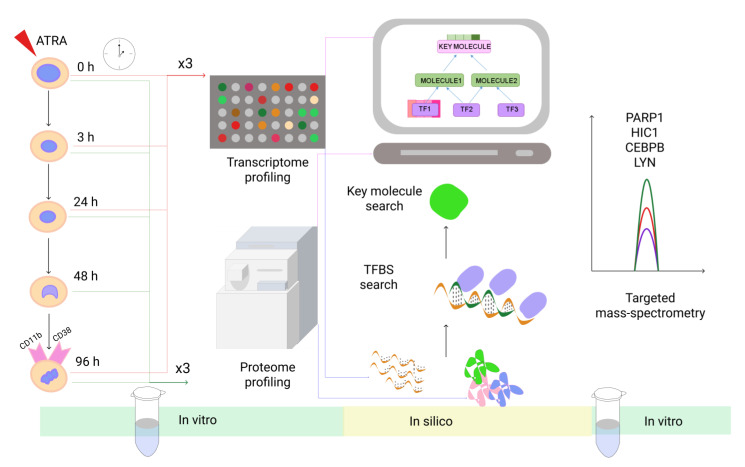
The study workflow. We applied a multi-disciplinary platform to study ATRA-induced granulocytic differentiation in a time-course manner using HL-60 cell line as a model. We combined LC-MS/MS analysis (0, 3, 24, 48, and 96 h after ATRA treatment, three bio repeats), whole-genome transcriptome analysis (0, 3, 24, and 96 h after ATRA treatment, three bio repeats), and bioinformatic search for transcription factor binding sites (TFBS) and for the key regulatory molecules. To verify the predicted regulatory networks the abundance of proteins HIC1, CEBPB, LYN, and PARP1, belonging to the designed model regulatory networks or involving in differentiation onset, were measured in time-course manner by selected reaction monitoring (SRM) using synthetic isotopically-labeled peptides as standard.