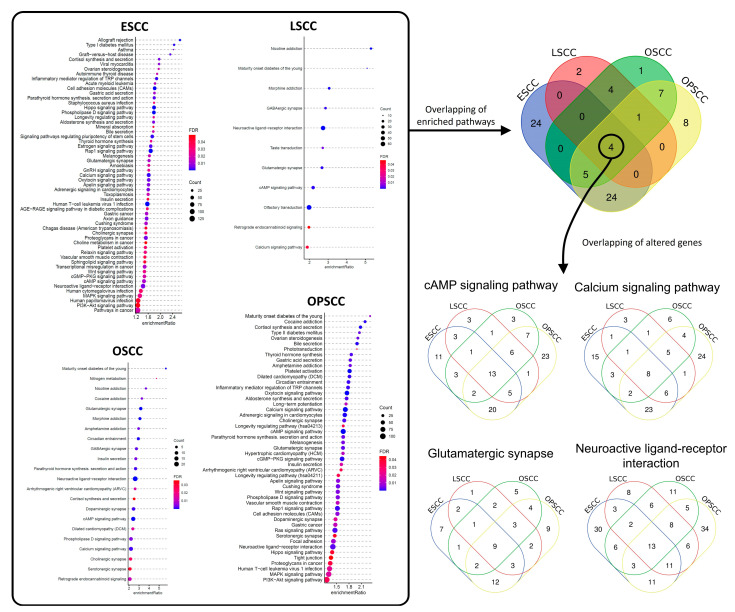
Figure 2.
Signaling pathway enrichment analysis with genes affected by differentially methylated regions. DMRs were identified by comparing each tumor type with its corresponding non-tumor tissue. In all comparisons, only DMRs containing at least 5 differentially methylated CpG sites, with FDR < 0.05 were considered. Genes with at least one DMR were included in the Over-representation analysis (ORA) using KEGG database to identify enriched signaling pathways. In the highlighted rectangle in the Figure, enriched pathways for each tumor type are shown (FDR < 0.05). The overlapping of enriched pathways (color filled Venn diagram) showed that 4 signaling pathways were over-represented in all tumor types, cAMP Signaling Pathway, Calcium Signaling Pathway, Glutamatergic Synapse, and Neuroactive Ligand-Receptor Interaction. However, the overlapping of DMR-carrying genes showed that (non-colored filled Venn diagrams) the genes affected varied in the different tumor types. ESCC, esophageal squamous cell carcinoma; LSCC, laryngeal squamous cell carcinoma; OSCC, oral cavity squamous cell carcinoma; OPSCC, oropharyngeal squamous cell carcinoma.