Abstract
The gastrointestinal tract is an important reservoir of extended spectrum beta-lactamase (ESBL)/carbapenemase-producing Enterobacterales isolates. This study included patients from two Bulgarian hospitals. Overall, 98 ESBL producers (including 68 Escherichia coli and 20 Klebsiella pneumoniae isolates) were detected among 99 hospitalized patients, 212 patients at admission, and 92 hospital staff in 42.4%, 24.5%, and 4%, respectively. We observed blaCTX-M-15 in 47% of isolates, blaCTX-M-3 in 39% and blaCTX-M-14 in 11%. Three blaCTX-M-15 positive isolates were also blaKPC-2 positive. High transferability was detected for blaCTX-M-3 carrying plasmids (55%) with L/M and I1 replicon plasmids, followed by CTX-M-14 (36.4%) and CTX-M-15 (27.9%) with IncF plasmids. BlaKPC-2 was carried by FIIAs plasmids. Epidemiology typing revealed 8 K. pneumoniae ST types—ST15(8/20), ST17(4/20), ST37(2/20) and 9 E. coli ST types—ST131 (30.9%, 21/68), ST38 (8/68), ST95(7/68) and ST316(7/68). All ST131 isolates but one was from the highly virulent epidemic clone O25bST131. This is the first report in Bulgaria about ESBL/carbapenemase faecal carriage. We observed high ESBL/carbapenemases prevalence. A predominant number of isolates were members of highly epidemic and virulent PanEuropean clones ST15 K. pneumoniae and O25bST131 E. coli. High antibiotics usage during the COVID pandemic will worsen the situation. Routine screenings and strict infection control measures should be widely implemented.
Keywords: fecal carriage, ESBL, carbapenemases, Enterobacterales
1. Introduction
Members of order Enterobacterales are Gram-negative bacteria normally inhabiting the intestinal tract. These bacteria are also the most common cause of nosocomial and community acquired infections [1,2,3]. The increasing rates of antibiotic resistance among them, especially in Escherichia coli, Klebsiella pneumoniae, and Enterobacter cloacae complex, have been globally reported [2,4,5]. The resistance was mostly associated with production of beta-lactamases (extended-spectrum beta-lactamases (ESBL) or/and carbapenemases) [2]. ESBLs could be classified into three main groups, TEM, SHV and CTX-M, with CTX-M-15 and CTX-M-14 being the most widespread variants [2]. Extensive usage of carbapenems for treatment of infections caused by ESBL producers has led to a sharp increase of carbapenemase producers (such as KPC, NDM, and VIM) [6]. Different mobile elements (plasmids and integrons) contribute to the dissemination of the producers. In the mobile elements, ESBL and carbapenemases genetic determinants are located with other resistance genes causing nonsusceptibility to beta-lactams and many other groups such as quinolones, aminoglycosides, tetracyclines, and trimethoprim/sulfamethoxazole [7].
The gastrointestinal tract is an important reservoir of ESBL/carbapenemase-producing enterobacteria [8]. It is also a hot spot where can occur exchanges of resistance genes and antibiotic pressure could result in appearance of resistant mutants [8]. This can lead to life-threatening nosocomial and community-acquired infections [8,9]. Fecal carriage of ESBL producers was widely reported all over the world [8,9]. In next decades such reports increased [8]. Wide differences in ESBL/carbapenemase fecal carriage rates have been reported in different countries, suggesting dynamics of their geographical evolution [8,10,11]. In Bulgaria, fecal carriage of ESBL/carbapenemase-producing Enterobacterales has not been investigated so far.
The aim of this study was to investigate the prevalence of ESBL and carbapenemase-producing Enterobacterales from fecal samples, collected from hospitalized patients (>48 h hospital-stay), and medical personnel in Varna hospital and from patients at admission in Sofia hospital, to discover the clonal relatedness between isolates and distribution of MLST types among E. coli (Achtman scheme) and K. pneumoniae (Pasteur scheme) isolates, as well as to characterize mechanisms of beta-lactam resistance.
2. Material and Methods
2.1. Bacterial Isolates
The study was conducted in two hospitals—University Hospital “Saint Marina”, Varna and Second City hospital in Sofia during the period January 2015–September 2015 (January–March 2015 for UH Varna and May-September 2015 for Sofia hospital). In University hospital in Varna, routine screening of 99 hospitalized patients in ICU words (>48 h after admission) was performed and in Sofia, 212 patients at the day of their admission were investigated. The fecal samples were sequentially collected, one per patient. They were inoculated on selective MacConkey agar with cefotaxime 1 mg/L and on ChromagarTMKPC (Becton Dickinson, Springfield, IL, USA). Bacterial isolates were identified using routine biochemical identification and were confirmed by VITEK (bioMérieux, Marcy L’Étoile, France)) or Phoenix (Becton Dickinson, Springfield, IL, USA). Antimicrobial susceptibility testing was performed using the disk diffusion method on Müller-Hinton II agar and the interpretation was by clinical breakpoints of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines (Version 10) (http://www.eucast.org/clinical_breakpoints/ accessed on 21 April 2021). The following antibiotics were tested: amoxicillin-clavulanic acid, piperacillin-tazobactam, cefotaxime, ceftazidime, cefepime, cefoxitin, imipenem, tobramycin, gentamicin, amikacin, trimethoprim/sulfamethoxazole, ciprofloxacin, and levofloxacin.
2.2. Phenotypic ESBL and Carbapenemases Detection
Presumptive ESBL production was detected with the double-disk synergy method [12]. In the case of nonsusceptibility to carbapenems or/and growth on selective ChromagarTMKPC media, a phenotypic confirmation of carbapenemase production was performed by the modified Hodge test and by the KPC/Metallo-beta-lactamase and OXA-48 Confirm Kit (ROSCO Diagnostica, Taastrup, Denmark). The beta-lactamase production was analyzed by isoelectric focusing (IEF) and the hydrolytic activity of individual beta-lactamase bands was assessed by a bioassay as previously described [13].
2.3. Molecular-Genetic Beta-Lactamase Identification
All isolates were screened for the presence of blaVIM, blaIMP, blaKPC, blaNDM, blaOXA-48, blaCTX-M, and blaSHV, as previously described [12,13,14,15]. The genes were sequenced using primers binding outside the coding region of blaSHV, blaCTX-M-1-group, blaKPC [12]. The nucleotide and deduced amino acid sequences were analyzed and multiple alignments were performed using Chromas Lite 2.01 (Technelysium Pty Ltd., Brisbane, Australia) and DNAMAN version 8.0 Software (Lynnon BioSoft, Vaudreuil-Dorion, QC, Canada).
2.4. Conjugation Experiments and Replicon Typing
Conjugation experiments were carried out as previously described [12]. Plasmid replicon typing were performed according to the Carattoli scheme [16]. E. coli phylotyping was done by Clermont scheme [17].
2.5. ERIC and MLST Typing
The clonal relatedness was investigated by ERIC PCR and Multilocus Sequence Typing (MLST). ERIC-1 and -2a primers were used [13]. Primers and protocols for E. coli MLST, Achtman scheme, were reported by Wirth et al. [18]. The assignment to allelic numbers and sequence types (STs) were performed according to the MLST database (https://bigsdb.web.pasteur.fr/ecoli/ecoli.html, accessed on 10 January 2021). Additionally, the pabB gene was detected to prove the presence of O25bST131 clone according to the Clermont scheme [19].
For the K. pneumoniae isolates, protocols and assignment to allelic numbers and sequence types (STs) were carried out as described in the MLST database (Pasteur Institute, Paris, France; http://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html accessed on 10 January 2021). A clonal complex was defined as a group of two or more independent isolates that shared six identical alleles.
3. Results
3.1. Bacterial Isolates
A total of 100 Enterobacterales isolates resistant to cefotaxime were isolated from the fecal samples (one per patient) of 99 patients, hospitalized in UH Varna (from ICU wards) and from the 212 patients, at the day of admission, in Second Town hospital (non-ICU wards)—Sofia during their admission. Ninety-two members of all medical personnel in ICU wards in “Saint Marina” hospital, Varna were studied. The obtained isolates were identified as Escherichia coli, n = 68; Klebsiella pneumoniae, n = 20; Enterobacter cloacae, n = 6; Klebsiella aerogenes (formerly Enterobacter aerogenes), n = 3, Shigella sonnei, n = 2, and Citrobacter freundii, n = 1.
3.2. Antimicrobial Susceptibility Testing
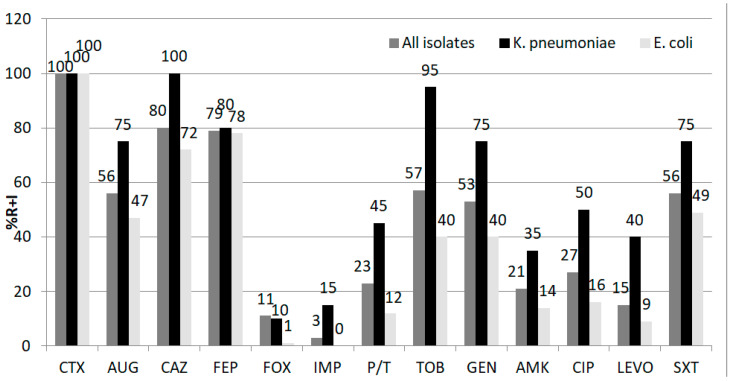
The nonsusceptibility rates (resistant and intermediately susceptible) of the whole group of isolates and separately for E. coli and K. pneumoniae are shown in Figure 1.
Figure 1.
Antimicrobial susceptibility rates in the whole group of Enterobacterales isolates (n = 98), E. coli (n = 68), and K. pneumoniae (n = 20). Abbreviations: CTX, cefotaxime; AUG, amoxicillin/clavulanic acid; CAZ, ceftazidime; FEP, cefepime; FOX, cefoxitine; IMP, imipenem; P/T, piperacillin/tazobactam; TOB, tobramycin, GEN, gentamycin; AMI, amikacin; CIP, ciprofloxacin; LEVO, levofloxacin; SXT, trimethoprim/sulfamethoxazole; R + I—resistant and intermediately susceptible isolates.
3.3. Phenotypic ESBL and Carbapenemases Detection
The ESBLs production was confirmed with double disk synergy test for 95 isolates, two E. cloacae isolates were possible AmpC hyperproducers (FOX-reistant and with antagonism shown when disk amoxicillin/clavulanic acid was placed near cefotaxime or ceftazidime). Three isolates resistant to imipenem gave positive modified Hodge test and increasing zone with disk meropenem+boronic acid suggesting class A carbapenemases activity.
3.4. Molecular-Genetic Beta-Lactamase Identification
The PCR and sequencing revealed the presence of blaESBL in 98 isolates and genes, encoding carbapenemases in 3 of them. The prevalence of (solely) ESBL-producing Enterobacterales was 30.2% (94 from 311) among patients (hospitalized and at admission) and 4% (4 from 92) among hospital staff. Three isolates from hospitalized patients (1%) produced carbapenemases as well. Among the hospitalized patients, the ESBL producer rate was 42.4% (42 of 99 patients) and among the patients at admission, they were 24.5% (52 of 212 patients). The difference was statistically significant (p = 0.0014).
We observed blaCTX-M-15 in 47% (46 from 98) of isolates and blaCTX-M-3 in 39% (38 from 98) (Table 1). Three blaCTX-M-15 positive isolates were also positive for blaKPC-2. According to the species, 41% (28/68) of E. coli and 70% (14/20) of K. pneumoniae were positive for blaCTX-M-15 (including KPC-2 producers); for blaCTX-M-3—40% of E. coli (27/68) and 30% of K. pneumoniae (6/20) were positive. Eleven (11%) isolates, all of them E. coli, showed the presence of blaCTX-M-14, two of blaSHV-12 and one of blaCTX-M-1.
Table 1.
Distribution of extended spectrum beta-lactamases (ESBLs)/carbapenemases according to species and patient’s type.
| ESBL. Species |
CTX-M-3 Number H/PA |
CTX-M-15 Number H/PA | KPC-2 + CTX-M-15 Number H/PA |
CTX-M-14 Number H/PA | CTX-M-1 Number H/PA | SHV-12 Number H/PA |
|---|---|---|---|---|---|---|
|
K. pneumoniae n = 20 |
4/2 | 11/0 | 3/0 | |||
| E. coli n = 68 | 9/18 | 6/22 | 0/1 | 8/3 | 0/1 | |
|
E. cloacae complex n = 5. |
1/0 | 2/1. | ||||
| K. aerogenes n = 2 | 2/0 | |||||
| C. freundii n = 1 | 1/0 | |||||
| S. sonnei n = 2 | 0/2 | |||||
| Total n = 98 | 16/22 | 20/23 | 3/0 | 0/1 | 8/3 | 1/1 |
Abbreviations: H—hospitalized patients, in bold, PA patients at admission.
3.5. Isoelectric Focusing and Bioassay
The isoelectric focusing was carried out in representative isolates for the species and the center and confirmed the beta-lactamase production of the ESBLs and carbapenemases. Of 20 K. pneumoniae isolates, 10 were subjected to IEF (three of the CTX-M-3-producing, four of the CTX-M-15-producing and all the KPC-2-producing isolates). We detected single band hydrolyzing cefotaxime with pI8.8 in blaCTX-M-15 positive, with pI8.4 in blaCTX-M-3 positive. For blaKPC-2 positive isolates two bands, hydrolyzing cefotaxime, were observed—with pI8.8 and with pI6,7. The band with pI 6.7 additionally hydrolyzed imipenem. Ten of the 27 blaCTX-M-3 E. coli isolates 11 from 22 blaCTX-M-15 positive and 6 from 11 blaCTX-M-14 positive isolates gave cefotaxime hydrolyzing band at pI8.8, pI8.4 and pI8.0, respectively. One E. coli isolate showed cefotaxime hydrolyzing band at pI8.2 (SHV-12) and another one at pI8.4 (CTX-M-1). All isolates have only one cefotaxime hydrolyzing band. There were not isolates with two ESBLs. Enterobacter isolates also produced only one cefotaxime hydrolyzing band.
3.6. Conjugation Experiments and Replicon Typing
Conjugation experiments showed 41 transconjugants. The most transferable gene was blaCTX-M-3 in 55% (23 of 38 donors), followed by blaCTX-M-14 in 36.4% (4 from 11) and blaCTX-M-15 in 27.9% (12 from 43 donors). We detected transfer of blaKPC-2 in two of three KPC-2-producing donors. The susceptibility phenotypes and replicon types of the transconjugants are shown in Table 2 and Table 3. blaCTX-M-3 was mainly associated with I1 (n = 11) and L/M (n = 10) replicons. Only single isolates showed FIIAs. blaCTX-M-15 was carried by IncF type (n = 8), HI2 (n = 1) and non-typeable plasmids (n = 3). FIIAs replicons were specific for blaKPC-2 carrying plasmids. Two isolates (1 CTX-M-3-producing E. cloacae complex and 1 K. aerogenes) gave transconjugants with L/M replicon type and CTX, TOB, GEN, AMI, SXT resistotype. The single C. freundii isolate gave CTX, TOB. GEN, CIP, TET, SXT transconjugant with HI2 replicon type.
Table 2.
ESBL/carbapenemases, Multilocus Sequence Typing (MLST), replicon typing, and transconjugant characteristics of 20 K. pneumoniae isolates.
| Patients, Type | ESBL | MLST Types | Phenotype of Transconjugants | Replicon Type |
|---|---|---|---|---|
| Hospitalized (14) | CTX-M-3 (4) | ST17(4) | CTX,TOB.GEN.AMI,SXT(2) | L/M(2) |
| CTX-M-15 (7) | ST15(5) * | - | ||
| ST340(1) ST16(1) |
- | |||
| KPC-2+CTX-M-15 (3) | ST15(3) | IMP,MER,PIP/TAZ (2) | FIA,FIIAs (2) | |
| At admission (6) | CTX-M-3 (2) | ST37(2) | CTX, GEN (2) | L/M(2) |
| CTX-M-15 (4) | ST215(1) ST359(1) ST873(2) |
CTX,CAZ,TOB,GEN,TET CTX,CAZ,TOB,GEN,CIP - |
F (1) NT (1) |
Abbreviations: CTX, cefotaxime; CAZ, ceftazidime; TOB, tobramycin, GEN, gentamycin; AMI, amikacin; TET, tetracycline, SXT, trimethoprim/sulfamethoxazole; * including two isolates from the medical personnel.
Table 3.
ESBLs, MLST, replicon typing and transconjugant characteristics of 68 E. coli isolates.
| ESBL | ERIC | MLST | Phylogroup | Phenotype of Transconjugants | Replicon Type | |
|---|---|---|---|---|---|---|
| Isolates from HP n = 23 |
CTX-M-3 n = 9 |
f(5) | ST131 * | A | CTX (2) | I1(2) |
| b(1) | ST 38 | D | CTX (1) | I1(1) | ||
| uni(3) | ND | A | CTX,TOB,GEN,AMI.SXT(1) CTX, SXT (1) |
L/M(1) I1(1) |
||
| CTX-M-14 n = 8 |
c(2) | ST 43 | A | - | ||
| a(4) | ST 95 | B2 | CTX(2) | F(2) | ||
| b(2) | ST 38 | D | - | |||
| CTX-M-15 n = 6 |
f(3) | ST131 | B2 | CTX,CAZ,TOB,GEN,TET(2) | FIA,F(2) | |
| d(2) | ST405 | D | CTX,CAZ,TOB,TET,SXT(1) | FIA,F(1) | ||
| l(1) | ST316 | A | - | |||
| Isoaltes from PA n = 45 |
SHV-12 n = 1 | f(1) | ST131 | B2 | - | |
| CTX-M-1 n = 1 | f(1) | ST131 | B2 | - | ||
| CTX-M-3 n = 18 |
f(5) ** | ST131 | B2 | CTX(2) CTX,TOB,GEN,AMI(1) CTX,TOB,GEN,SXT,CHL(1) |
I1(2) L/M(1) FIIAs(1) |
|
| l(3) | ST316 | A | CTX(1) | I1(1) | ||
| b(2) | ST 38 | D | CTX(1) | I1(1) | ||
| m(2) | ST676 | B2 | CTX,GEN(1) | L/M(1) | ||
| a(1) | ST 95 | B2 | CTX(1) | I1(1) | ||
| t(3) | ST 517 | B1 | CTX,GEN(1) | L/M(1) | ||
| uni(2) | ND | A(2) | CTX(2) | I1(2) | ||
| CTX-M-14 n = 3 |
a(1) | ST 95 | B2 | CTX(1) | F(1) | |
| f(1) | ST131 | B2 | CTX(1) | F(1) | ||
| m(1) | ST676 | B2 | ||||
| CTX-M-15 n = 22 |
f(5) | ST131 | B2 | |||
| l(3) | ST316 | A | CTX,CAZ(2) CTX,CAZ,SXT(1) |
F(3) | ||
| a(1) | ST 95 | B2 | ||||
| m(2) | ST676 | B2 | CTX,CAZ(1) | F(1) | ||
| n(2) | ST540 | A | CTX,CAZ(1) | NT(1) | ||
| b(3) | ST38 | D | CTX,CAZ(1) | NT(1) | ||
| uni (6) | ND | A(5),B1(1) |
Abbreviations: CTX, cefotaxime; CAZ, ceftazidime; TOB, tobramycin, GEN, gentamycin; AMI, amikacin; TET, tetracycline, SXT, trimethoprim/sulfamethoxazole; ND—not determined, * including two isolates from the medical personnel; ** including one pab negative isolate; HP, hospitalized patients; PA, patients at admission.
3.7. MLST and ERIC Typing
Epidemiology typing revealed 8 ERIC clusters in K. pneumoniae isolates, they had between 1 and 7 members, which corresponded to 8 ST types—ST15(8 isolates, 40%), ST17(4 isolates), ST37(2 isolates), ST873(2 isolates) and ST215, ST359, ST340, ST16 as single isolates (Table 2). ST15 K. pneumoniae isolates were prevalent among hospitalized patients and were associated with CTX-M-15 and KPC-2 production (Table 1 and Table 2). In this clone, two K. pneumoniae CTX-M-15-producing isolates from the medical staff were also included.
Among E. coli isolates, 20 ERIC types were detected, nine of them had between 2 and 21 members. They corresponded to 9 main ST types (Table 3)—ST131n=21 (type f) (30.9%), ST38n=8 (type b), ST95n=7 (type a), ST316n=7 (type l), ST43n=2 (type c), ST540n=2 (type n), ST676n=5 (type m), ST405 n=2 (type d), ST517n=3 (type t). Eleven isolates represented unique ERIC types and MLST have not been performed. All ST131 E. coli isolates but one were positive for pabB gene.
ST131 E. coli (n = 21) was detected predominantly among patients at admission and was less prevalent among hospitalized patients, and was associated with production of CTX-M-15 (38.1%, 8/21), CTX-M-3 (47.6%, 10/21) and CTX-M-1, CTX-M-14 and SHV-12 as single isolates.
The following E. coli phylotypes were identified: B2n=33 (48.5%, 33/68), An=21, B1n=4, Dn=10. The association between MLST types and phylotypes is shown in Table 3. The B2 group included members of ST131, ST95, ST 676, D group—members of ST38 and ST405, A group—ST316, ST43, almost all the unique ERIC types and B1—ST517.
With exception of two CTX-M-15-producing E. cloacae isolates, all demonstrated unique ERIC types S. sonnei generated only three bands in ERIC PCR, so, they were not enough to compare the isolates.
4. Discussion
To the best of our knowledge, this is the first report on intestinal carriers of ESBL/carbapenemase-producing Enterobacteriales isolates in Bulgaria. This study revealed a high (30.2%) rate of fecal ESBL producers (including 3% carbapenemase producers) among 311 Bulgarian patients. Importantly, the rate of ESBL-producing strains among hospitalized patients (99 patients) was statistically higher compared with that in patients at admission (212 patients) (42.4% vs. 25.5%). The ESBL prevalence among patients at admission could reflect the frequency of ESBL producers in the community and it was higher than that in other European countries [10]. Our results were higher than those in Portuguese hospitals, where authors compared ESBL frequency among hospitalized, >48 h hospital stay (24%) and patients at admission (17%) [20]. Similar results have been reported in French hospitals—with 17.7% ESBL gut carriage [21]. Our rates were much higher than those published from other European countries (for patients at admission) such as Belgium—11.6% [22], UK—9.5% [23], Spain—7.7% [24] and The Netherlands—8.2% [25]. All these data were in concordance with a very high level of cephalosporin 3rd resistant invasive isolates in Bulgaria. In 2019 ECDC reports the highest frequency of resistance to cephalosporin third generation in invasive isolates of E. coli (38.6%), and K. pneumoniae from Bulgaria (75.7%). Unfortunately, the frequency of these isolates in Bulgaria was the highest in Europe and has remained high over the last five years (https://www.ecdc.europa.eu/sites/default/files/documents/Additional-tables-EUEEA-population-weighted-mean-2019.pdf, accessed on 23 April 2021). Many reports focused on increasing rates of gut colonization with ESBL/carbapenemase producers among patients infected by ESBL/carbapenemase-producing microorganisms [26]. The high frequency of ESBL intestinal carriage is a threat of an increase in infections caused by ESBL producers. The rate of ESBL gut carriers in our study was much lower among the medical personnel (4%), but their appearance showed that medical staff can also contribute to the dissemination of these isolates.
A reason for the high frequency of fecal ESBL-producing Enterobacteriales isolates in the present study and cephalosporin third generation resistant invasive isolates in Bulgaria could be a high level of cephalosporin and carbapenems consumption in our country. Bulgaria was the first place of usage of third generation cephalosporins in Europe with 57% of the total antimicrobial consumption and in last place for penicillin usage (7%) in 2019 (https://www.ecdc.europa.eu/sites/default/files/documents/Antimicrobial-consumption-in-the-EU-Annual-Epidemiological-Report-2019.pdf, accessed on 23 April 2021). Strict measures for infection control and good compliance with antibiotic policy are necessary.
Our isolates had the following resistance rates to non—beta-lactam antimicrobials: 21–57% for aminoglycosides and 15–27% for quinolones. The resistance rates for these agents were much higher in Klebsiella than in E. coli isolates (Figure 1). Only three isolates from the studied group (K. pneumoniae) were carbapenem-resistant. Carbapenems, amikacin, and levofloxacin are adequate options for empirical antimicrobial therapy when necessary. Piperacillin/tazobactam can also be considered after antimicrobial susceptibility testing. Monitoring of the patients colonized by ESBL/carbapenemases producers should be regularly performed.
As in other reports [8,10], we detected the prevalence of E. coli ESBL-producing fecal isolates (69%), and in less proportion Klebsiella ESBL-producing isolates (20.4%). Similar to other studies we found a predominance of CTX-M-1 group producers [8,10]. The CTX-M-15 in the present study was predominantly found among K. pneumoniae and E. cloacae complex isolates (Table 1). E. coli isolates produced CTX-M-15 and CTX-M-3 in almost equal proportions, in 41% and 40%, respectively. We could assume that blaCTX-M-15 allele might offer the bacteria a selective advantage to overcome antibiotic pressure in hospitals. CTX-M-14 enzyme has been previously detected in Bulgaria only sporadically [27]. Interestingly, in this study, we observed it among E. coli and in only 16%. Gut colonization with CTX-M-15 producers has been reported elsewhere [8,10] (including gut carriers), whereas CTX-M-9 and CTX-M-14 from the same origin have been reported in few countries—Spain and China [8].
Only three isolates K. pneumoniae (3%) produced KPC-2 carbapenemases and they were in combination with CTX-M-15 production. In Europe, the rates of carbapenemase producers has increased during the last years [5,6] and one important contributor for this is intestinal carriage of carbapenemases producers. The presence of carbapenemase carriers is of high importance since carbapenemase producers could not be easily cleared from the body. The process can take several months (>387 days) [28].
In the present study, ST15 was the main MLST type among K. pneumoniae and was associated with CTX-M-15 and KPC-2 production. The ST15 clone is an interesting PanEuropean clone reported to produce wide range of ESBL and carbapenemases [29,30,31,32,33]. Among the hospitalized patients, this clone could be an important driver of KPC-2 production and/or CTX-M-15 production. In the present study, 8 of 20 investigated K pneumoniae isolates were from this clone and three of them produced KPC-2. It is also very important that two isolates from the medical staff had CTX-M-15-producing ST15 K. pneumoniae. In Bulgaria, during the period 2012–2015, clinical KPC-2-producing ST15 K. pneumoniae isolates were detected in Varna hospital [13]. Our data shows the great transmissible potential of ST15 isolates including those from gut colonization. This comes to show that the gut microflora can act as a reservoir of carbapenemase producers, therefore, screening for fecal carriage is important, especially for immunosuppressed and transplanted patients.
The other intestinal ST K. pneumoniae types in this study were ST17 and ST37. They produced CTX-M-3 enzymes among hospitalized patients. These STs have also been isolated previously from clinical isolates in Bulgaria [34], but were mostly associated with CTX-M-15 production. We could suggest that additionally to clonal propagation, plasmid distribution plays an important role for ESBL determinants dissemination. Similar to other reports in our study blaCTX-M-3 was predominantly carried by IncL/M plasmid and IncI1 [7]. In the present study, K. pneumoniae from in admission patients showed the presence of rare ST types, possibly reflecting the isolates in the community.
Among E. coli isolates, we observed 9 ST types of the most common ST131(30.9%), ST38(11.8%), ST95(10.3%), ST316 and ST405. Interestingly, all isolates from ST131 were of phylogroup B2 and all but one were of O25bST131 clone. Our findings showed that O25bST131 represented the major clone among fecal E. coli isolates harboring different ESBL genes—CTX-M-15, CTX-M-3, CTX-M-14, CTX-M-1, and SHV-12. Similar to results of other authors, we have observed it mainly among patients at admission, showing that the community could be a reservoir for this clone [35,36]. ST131 was more likely to be transmitted between members of the same family than within patients in the hospital environment [37]
The fact that O25bST131 members were detected also among medical personnel is alarming. Strict infection control measures and education of personnel were applied and six months later, ESBL-producing isolates were no more found among the medical staff (personal communication). O25bST131 is a widely distributed, highly virulent, and epidemic PanEuropean clone, that could easily be transmitted all over the world and is associated with CTX-M-15, OXA-1, and quinolone resistance. Our results showed the potential of this clone to produce a wide range of enzymes—in addition to CTX-M-15, also CTX-M-3, CTX-M -14, CTX-M-1, and SHV-12. Its virulence enables the bacteria to cause both hospital- and community acquired infections [38]. Some authors report an association of O25bST131 with prolonged gut carriage [39].
Our isolates from the ST95 (B2 phylogroup) clone were mainly associated with CTX-M-14 production, five of seven ST95 isolates produced CTX-M-14. This clone has been referred to as an important clinical clone associated with urinary and bloodstream infection [36,40,41]. ST38, belonged to D phylogroup, also has been commonly reported in clinical E. coli isolates and reported as a high-risk clone [4]. In this study, ST38 takes the second place after ST131 according to its frequency and was found to produce CTX-M-3, CTX-M-14 and CTX-M-15 enzymes.
In our study, two isolates of E. coli ST405 produced CTX-M-15. This clone was previously detected in Bulgaria as a carrier of NDM-1 carbapenemases [42]. E coli ST405 is an emerging urosepsis pathogen, reported to carry bla CTX-M, bla NDM, and a number of virulence genes comparable with O25b:H4-ST131 [43].
E. coli phylogroups B1 and A have been reported as commensal gut isolates and B2 and D to have pathogenic potential [4]. In the present study, the phylotyping showed that isolates members of B2 and D phylogroups were prevalent. ST 38, ST 95 and ST405 belonged to more virulent B2 and D groups and have been associated with increased virulence [4].
Among CTX-M-3 carriers, two types of plasmids were detected. IncL/M plasmid has been responsible for transmission among CTX-M-3-producing K. pneumoniae, Enterobacter spp. and S. sonnei isolates. In addition to IncL/M, among E. coli IncI1 has been confirmed too. CTX-M-15 production was associated with IncF and HI2 plasmids. The transconjugants showed resistance not only to cephalosporins third generation but also to aminoglycosides and co-trimoxazole. These antimicrobials could also make selective pressure. The high number of the isolated E. coli ST types showed that in addition to the plasmid, clonal transmission also plays an important role in distribution of ESBL genes. BlaKPC-2 were strongly associated with FIIAs replicon plasmids, as was reported previously [13].
In conclusion, this is the first report in Bulgaria about gut colonization with ESBL and/or carbapenemase producers. A high frequency of ESBL gut producers among hospitalized patients (42.4%) and in patients at admission (25.5%) was found. Three percent of isolates were KPC-2-producing K. pneumoniae. Only 4% of the medical staff evaluated carried ESBLs. The prevailing number of isolates were members of two highly epidemic and virulent PanEuropean clones, ST15 K. pneumoniae (40%, including two isolates from the medical staff) and ST131 clone (31%, also including 2 isolates from the medical staff). The isolates produced a wide range of ESBLs—CTX-M-15 (47%), CTX-M-3 (39%), and CTX-M-14 (11%).
In the present COVID pandemic situation, the antimicrobial overconsumption will increase the selective pressure both in the hospitals and in the community and may increase the rate and dissemination of ESBL/carbapenemase producers. Further studies should closely monitor this potential negative trend. Routine screening at hospital admission and during the hospital stay, as well as strict infection control measures should be widely implemented. In the present situation, adequate antimicrobial stewardship strategies are of critical importance for the restriction of unnecessary antimicrobial usage and prevention of the wide dissemination of problematic MDR bacteria.
Author Contributions
Conceptualization, R.M. and L.B.; methodology, R.M., P.S., D.I.; formal analysis, P.S., R.M., D.I.; Investigation, R.M., P.S., T.S., D.P., R.K., D.I.; Resourses Writing-original draft preparation, R.M.; Review and editing, R.M., L.B., T.S.; Supervision, R.M.; Project administration, R.M., L.B.; Funding acquisition, R.M.; Final reading and approval R.M., P.S., T.S., D.P.,D.I. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Medical University—Sofia, Bulgaria, grant №71/23.04.2019, project № 8242/20.11.2018 (RM).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Davin-Regli A., Lavigne J.P., Pagès J.M. Enterobacter spp.: Update on Taxonomy, Clinical Aspects, and Emerging Antimicrobial Resistance. Clin. Microbiol. Rev. 2019;32:e00002-19. doi: 10.1128/CMR.00002-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bush K., Patricia A. Bradford Epidemiology of β-Lactamase-Producing Pathogens. Clin. Microbiol. Rev. 2020;33:e00047-19. doi: 10.1128/CMR.00047-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wyres K.L., Lam M.M.C., Holt K.E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020;18:344–359. doi: 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 4.Peirano G., Pitout J. Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae: Update on Molecular Epidemiology and Treatment Options. Drugs. 2019;79:1529–1541. doi: 10.1007/s40265-019-01180-3. [DOI] [PubMed] [Google Scholar]
- 5.Bonomo R.A. β-Lactamases: A Focus on Current Challenges. Cold Spring Harb. Perspect. Med. 2017;7:a025239. doi: 10.1101/cshperspect.a025239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nordmann P., Poirel L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin. Infect. Dis. 2019;69(Suppl. 7):S521–S528. doi: 10.1093/cid/ciz824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Partridge S.R., Kwong S.M., Firth N., Jensen S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018;31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woerther P.L., Burdet C., Chachaty E., Andremont A. Trends in human fecal carriage of extended-spectrum β-lactamases in the community: Toward the globalization of CTX-M. Clin. Microbiol. Rev. 2013;26:744–758. doi: 10.1128/CMR.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thaden J.T., Fowler V.G., Sexton D.J., Anderson D.J. Increasing Incidence of Extended-Spectrum β-Lactamase-Producing Escherichia coli in Community Hospitals throughout the Southeastern United States. Infect. Control Hosp. Epidemiol. 2016;37:49–54. doi: 10.1017/ice.2015.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karanika S., Karantanos T., Arvanitis M., Grigoras C., Mylonakis E. Fecal Colonization With Extended-spectrum Beta-lactamase-Producing Enterobacteriaceae and Risk Factors among Healthy Individuals: A Systematic Review and Metaanalysis. Clin. Infect. Dis. 2016;63:310–318. doi: 10.1093/cid/ciw283. [DOI] [PubMed] [Google Scholar]
- 11.Jolivet S., Vaillant L., Poncin T., Lolom I., Gaudonnet Y., Rondinaud E., Bendjelloul G., Lomont A., Lucet J.C., Armand-Lefèvre L. Prevalence of carriage of extended-spectrum β-lactamase-producing enterobacteria and associated factors in a French hospital. Clin. Microbiol. Infect. 2018;24:1311–1314. doi: 10.1016/j.cmi.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Jarlier V., Nicolas M.H., Fournier G., Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: Hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 1988;10:867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 13.Markovska R., Stoeva T., Schneider I., Boyanova L., Popova V., Dacheva D., Kaneva R., Bauernfeind A., Mitev V., Mitov I. Clonal dissemination of multilocus sequence type ST15 KPC-2-producing Klebsiella pneumoniae in Bulgaria. APMIS. 2015;123:887–894. doi: 10.1111/apm.12433. [DOI] [PubMed] [Google Scholar]
- 14.Poirel L., Walsh T.R., Cuvillier V., Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 2011;70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Rasheed J.K., Kitchel B., Zhu W., Anderson K.F., Clark N.C., Ferraro M.J., Savard P., Humphries R.M., Kallen A.J., Limbago B.M. New Delhi metallo-β-lactamase-producing Enterobacteriaceae, United States. Emerg. Infect. Dis. 2013;19:870–878. doi: 10.3201/eid1906.121515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K.L., Threlfall E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods. 2005;63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 17.Clermont O., Bonacorsi S., Bingen E. Rapid and simple detection of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000;66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wirth T., Falush D., Lan R., Colles F., Mensa P., Wieler L.H., Karch H., Reeves P.R., Maiden M.C., Ochman H., et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clermont O., Dhanji H., Upton M., Gibreel T., Fox A., Boyd D., Mulvey M.R., Nordmann P., Ruppé E., Sarthou J.L., et al. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J. Antimicrob. Chemother. 2009;64:274–277. doi: 10.1093/jac/dkp194. [DOI] [PubMed] [Google Scholar]
- 20.Aires-de-Sousa M., Lopes E., Gonçalves M.L., Pereira A.L., Machado E Costa A., de Lencastre H., Poirel L. Intestinal carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae at admission in a Portuguese hospital. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:783–790. doi: 10.1007/s10096-019-03798-3. [DOI] [PubMed] [Google Scholar]
- 21.Pilmis B., Cattoir V., Lecointe D., Limelette A., Grall I., Mizrahi A., Marcade G., Poilane I., Guillard T., Bourgeois Nicolaos N., et al. Carriage of ESBL-producing Enterobacteriaceae in French hospitals: The PORTABLSE study. J. Hosp. Infect. 2018;98:247–252. doi: 10.1016/j.jhin.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 22.Schoevaerdts D., Verroken A., Huang T.D., Frennet M., Berhin C., Jamart J., Bogaerts P., Swine C., Glupczynski Y. Multidrug-resistant bacteria colonization amongst patients newly admitted to a geriatric unit: A prospective cohort study. J. Infect. 2012;65:109–118. doi: 10.1016/j.jinf.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Otter J.A., Natale A., Batra R., Tosas Auguet O., Dyakova E., Goldenberg S.D., Edgeworth J.D. Individual- and community-level risk factors for ESBL Enterobacteriaceae colonization identified by universal admission screening in London. Clin. Microbiol. Infect. 2019;25:1259–1265. doi: 10.1016/j.cmi.2019.02.026. [DOI] [PubMed] [Google Scholar]
- 24.Díaz-Agero Pérez C., López-Fresneña N., Rincon Carlavilla A.L., Hernandez Garcia M., Ruiz-Garbajosa P., Aranaz-Andrés J.M., Maechler F., Gastmeier P., Bonten M.J.M., Canton R. Local prevalence of extended-spectrum beta-lactamase (ESBL) producing Enterobacteriaceae intestinal carriers at admission and co-expression of ESBL and OXA-48 carbapenemase in Klebsiella pneumoniae: A prevalence survey in a Spanish University Hospital. BMJ Open. 2019;9:e024879. doi: 10.1136/bmjopen-2018-024879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Platteel T.N., Leverstein-van Hall M.A., Cohen Stuart J.W., Thijsen S.F., Mascini E.M., van Hees B.C., Scharringa J., Fluit A.C., Bonten M.J. Predicting carriage with extended-spectrum beta-lactamase-producing bacteria at hospital admission: A cross-sectional study. Clin. Microbiol. Infect. 2015;21:141–146. doi: 10.1016/j.cmi.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 26.Titelman E., Hasan C.M., Iversen A., Naucler P., Kais M., Kalin M., Giske C.G. Faecal carriage of extended-spec-trum beta-lactamase-producing Enterobacteriaceae is common 12 months after infection and is related to strain factors. Clin. Microbiol. Infect. 2014;20:O508–O515. doi: 10.1111/1469-0691.12559. [DOI] [PubMed] [Google Scholar]
- 27.Markovska R., Schneider I., Ivanova D., Mitov I., Bauernfeind A. Predominance of IncL/M and IncF plasmid types among CTX-M-ESBL-producing Escherichia coli and Klebsiella pneumoniae in Bulgarian hospitals. APMIS. 2014;122:608–615. doi: 10.1111/apm.12204. [DOI] [PubMed] [Google Scholar]
- 28.Zimmerman F.S., Assous M.V., Bdolah-Abram T., Lachish T., Yinnon A.M., Wiener-Well Y. Duration of carriage of carbapenem-resistant Enterobacteriaceae following hospital discharge. Am. J. Infect. Control. 2013;41:190–194. doi: 10.1016/j.ajic.2012.09.020. [DOI] [PubMed] [Google Scholar]
- 29.Rodrigues C., Machado E., Ramos H., Peixe L., Novais Â. Expansion of ESBL-producing Klebsiella pneumoniae in hospitalized patients: A successful story of international clones (ST15, ST147, ST336) and epidemic plasmids (IncR, IncFIIK) Int. J. Med. Microbiol. 2014;304:1100–1108. doi: 10.1016/j.ijmm.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Wang J.T., Wu U.I., Lauderdale T.L., Chen M.C., Li S.Y., Hsu L.Y., Chang S.C. Carbapenem-nonsusceptible Enterobacteriaceae in Taiwan. PLoS ONE. 2015;10:e0121668. doi: 10.1371/journal.pone.0121668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esteban-Cantos A., Aracil B., Bautista V., Ortega A., Lara N., Saez D., Fernández-Romero S., Pérez-Vázquez M., Navarro F., Grundmann H., et al. The Carbapenemase-Producing Klebsiella pneumoniae Population Is Distinct and More Clonal than the Carbapenem-Susceptible Population. Antimicrob. Agents Chemother. 2017;61:e02520-16. doi: 10.1128/AAC.02520-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ríos E., López M.C., Rodríguez-Avial I., Culebras E., Picazo J.J. Detection of Escherichia coli ST131 clonal complex (ST705) and Klebsiella pneumoniae ST15 among faecal carriage of extended-spectrum β-lactamase- and carbapenemase-producing Enterobacteriaceae. J. Med. Microbiol. 2017;66:169–174. doi: 10.1099/jmm.0.000399. [DOI] [PubMed] [Google Scholar]
- 33.Markovska R., Stoeva T., Boyanova L., Stankova P., Schneider I., Keuleyan E., Mihova K., Murdjeva M., Sredkova M., Lesseva M., et al. Multicentre investigation of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in Bulgarian hospitals—Interregional spread of ST11 NDM-1-producing K. pneumoniae. Infect. Genet. Evol. 2019;69:61–67. doi: 10.1016/j.meegid.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 34.Markovska R., Stoeva T., Boyanova L., Stankova P., Pencheva D., Keuleyan E., Murjeva M., Sredkova M., Ivanova D., Lazarova G., et al. Dissemination of successful international clone ST15 and clonal complex 17 among Bulgarian CTX-M-15 producing K. pneumoniae isolates. Diagn. Microbiol. Infect. Dis. 2017;89:310–313. doi: 10.1016/j.diagmicrobio.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 35.Banerjee R., Johnston B., Lohse C., Porter S.B., Clabots C., Johnson J.R. Escherichia coli sequence type 131 is a dominant, antimicrobial-resistant clonal group associated with healthcare and elderly hosts. Infect. Control Hosp. Epidemiol. 2013;34:361–369. doi: 10.1086/669865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathers A.J., Peirano G., Pitout J.D. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 2015;28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hilty M., Betsch B.Y., Bogli-Stuber K., Heiniger N., Stadler M., Kuffer M., Kronenberg A., Rohrer C., Aebi S., Endimiani A., et al. Transmission dynamics of extended-spectrum ß-lactamase-producing Enterobacteriaceae in the tertiary care hospital and the household setting. Clin. Infect. Dis. 2012;55:967–975. doi: 10.1093/cid/cis581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogers B.A., Sidjabat H.E., Paterson D.L. Escherichia coli O25b-ST131: A pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 2011;66:1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 39.Overdevest I., Haverkate M., Veenemans J., Hendriks Y., Verhulst C., Mulders A., Couprie W., Bootsma M., Johnson J., Kluytmans J. Prolonged colonisation with Escherichia coli O25:ST131 versus other extended-spectrum beta-lactamase-producing E. coli in a long-term care facility with high endemic level of rectal colonisation, the Netherlands, 2013 to 2014. Euro Surveill. 2016;21:30376. doi: 10.2807/1560-7917.ES.2016.21.42.30376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doumith M., Day M., Ciesielczuk H., Hope R., Underwood A., Reynolds R., Wain J., Livermore D., Woodford N. Rapid identification of major Escherichia coli sequence types causing urinary tract and bloodstream infections. J. Clin. Microbiol. 2015;53:160–166. doi: 10.1128/JCM.02562-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hertz F., Nielsen J., Schønning K., Littauer P., Knudsen J., Løbner-Olesen A., Frimodt-Møller N. Population structure of drug-susceptible,-resistant and ESBL-producing Escherichia coli from community-acquired urinary tract. BMC Microbiol. 2016;16:63. doi: 10.1186/s12866-016-0681-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poirel L., Savov E., Nazli A., Trifonova A., Todorova I., Gergova I., Nordmann P. Outbreak caused by NDM-1- and RmtB-producing Escherichia coli in Bulgaria. Antimicrob. Agents Chemother. 2014;58:2472–2474. doi: 10.1128/AAC.02571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy Chowdhury P., McKinnon J., Liu M., Djordjevic S.P. Multidrug Resistant Uropathogenic Escherichia coli ST405 With a Novel, Composite IS26 Transposon in a Unique Chromosomal Location. Front. Microbiol. 2019;9:3212. doi: 10.3389/fmicb.2018.03212. [DOI] [PMC free article] [PubMed] [Google Scholar]