Figure 1.
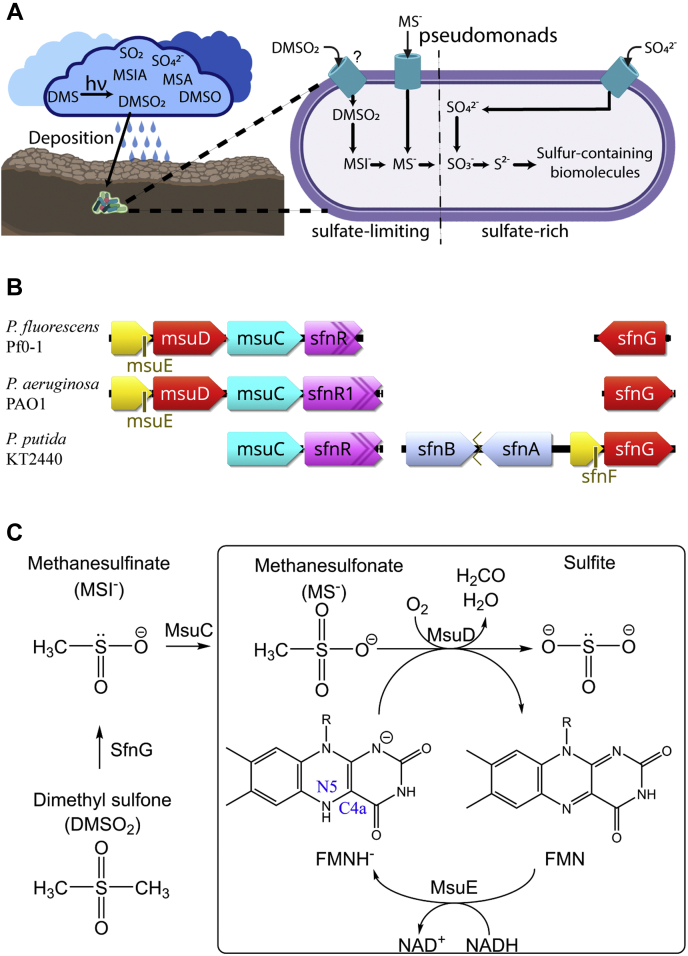
Bacterial sulfur assimilation of dimethyl sulfone and methanesulfonate.A, the cycling of atmospheric sulfur is depicted. Oxidation products of dimethylsulfide (DMS) include dimethyl sulfoxide (DMSO), dimethyl sulfone (DMSO2), methanesulfinic acid (MSIA), methanesulfonic acid (MSA), sulfur dioxide (SO2), and sulfate (SO42−). Pseudomonads can intracellularly convert DMSO2 into methanesulfinate (MSI−), methanesulfonate (MS−), and, finally, sulfite. Transporters for DMSO2 have yet to be characterized. Under sulfate-rich conditions, sulfate is reduced to sulfite and subsequently sulfide (S2−) for biosynthetic reactions. Representation created with BioRender.com. B, representative gene clusters for utilization of DMSO2 from P. fluorescens Pf0-1, P. aeruginosa PAO1, and P. putida KT2440 include three monooxygenases, sfnG, msuC, and msuD, and the oxidoreductases msuE or sfnF. Monooxygenases that break C–S bonds are in red, monooxygenases that form S–O bonds are in cyan, NADH:FMN oxidoreductases are in yellow, transcriptional regulators are in pink, and proteins of unknown function are in light blue. Initial studies of the sfn operon were reported for the genetically intractable P. putida DS1 (16, 29); therefore, P. putida KT2440 was used for alignment. The sfn operon in P. putida contains the uncharacterized sfnAB pair (79), which is separated by predicted ABC transporters and a hypothetical protein (represented as a jagged line); however, sfnG is a stand-alone gene within P. aeruginosa PAO1 (80) and P. fluorescens Pf0-1. The msu operon groups with sfnR, or sfnR1 and sfnR2 in P. aeruginosa (sfnR2 omitted from alignment) (80). Gene information is in Table S1. C, the sulfur assimilation pathway of P. fluorescens converts DMSO2 into sulfite. The NADH:FMN oxidoreductase MsuE is required to produce FMNH− for SfnG, MsuC, and MsuD; however, MsuE is shown only for the MsuD reaction for clarity.