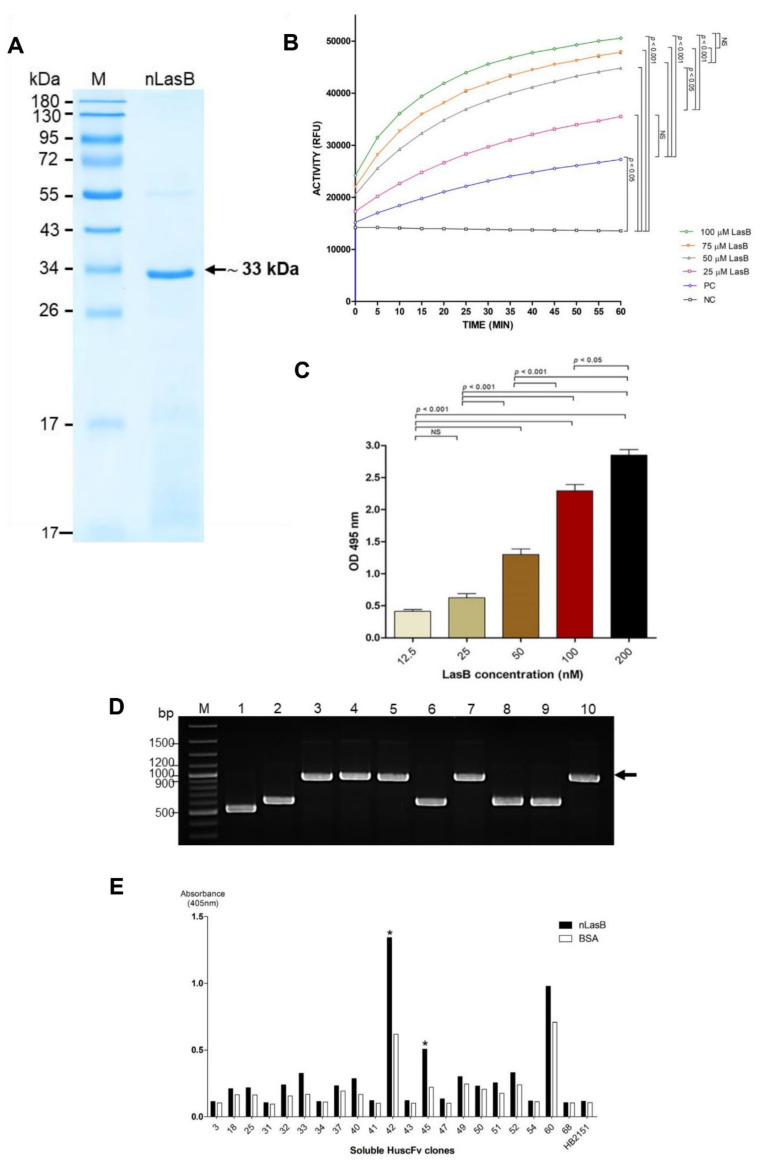
Figure 1.
Preparation of native LasB (nLasB) and selection of the LasB-bound HuscFv-displaying phage clones. (A) Purified nLasB revealed by SDS-PAGE and CBB staining. M, protein molecular mass marker (ThermoFisher Scientific, Rockford, lL, USA); nLasB, purified native LasB (~33 kDa, arrow). Numbers at the left are protein masses in kDa. (B) Determination of enzymatic activities of the purified nLasB using fluorogenic substrate assay. PC, positive control which was porcine pancreatic elastase (0.025 U/mL) mixed with substrate; NC, negative control which was reaction buffer mixed with substrate; ACTIVITY (RFU), relative fluorescence units of the enzymatic activity per minute. (C) Results of elastin-Congo Red assay for determining elastolytic activity of the nLasB. NS, not significantly different. (D) Colony-PCR analysis of representative phage-transformed HB2151 E. coli clones; black arrow indicates amplicon of complete huscfvs (1000 bp) in the phage-transformed-E. coli. Smaller DNA bands are truncated huscfvs. M, DNA marker (Thermo Fisher Scientific); 1-10, the phage-transformed E. coli clones nos. 1-10, respectively. Numbers at the left are DNA masses in base pairs (bp). (E) Results of indirect ELISA for determining the binding of HuscFvs in lysates of phage-transformed HB2151 E. coli clones to nLasB, control BSA (antigen control), and HB2151 (lysate of original HB2151 E. coli without huscfv-phagemid) as background binding control; asterisks indicate the clones that their HuscFvs gave the high binding activity to nLasB (an ELISA signal at OD 405 nm to the nLasB: OD 405 nm to the control BSA was more than 2). These clones were selected for further experiments.