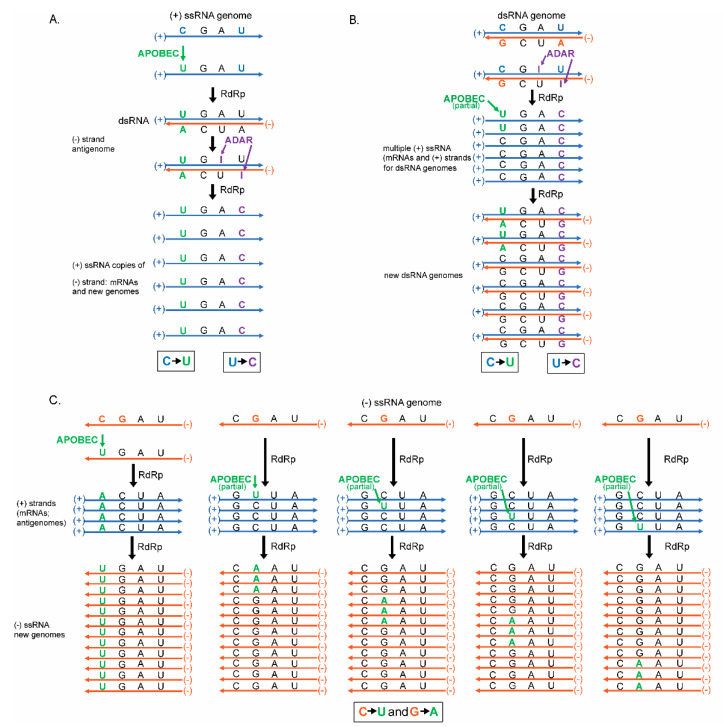
Figure 1.
RNA virus genome type and mode of replication define mutation strand bias in the progeny. Presented are the modes of RNA viral genome replication and how mutagenesis with the ssRNA-specific cytidine deaminase APOBEC and the dsRNA-specific adenosine deaminase ADAR affect the genomes of the cell. Positive (+) strands are shown in blue. Negative (−) strands are shown in orange. Color codes, same as a strand color, are assigned to nucleotides that will be mutated in the next steps. APOBEC mutagenesis and resulting mutant nucleotides are shown in green. ADAR mutagenesis in dsRNA and resulting mutant nucleotides are shown in purple. Nucleotides that stayed not mutated in the progeny are shown in black. Predominant classes of mutations in progeny ssRNA genomes or in coding (+) strand RNA of dsRNA genomes is shown in boxes. (A) Viruses with positive (+) ssRNA genome. The infecting (+) strand RNA genome is used as a template by RNA dependent RNA polymerase (RdRp) to synthesize a dsRNA with both (+) and (−) strands. A single dsRNA molecule is subsequently used to generate multiple copies of (+) strand RNA transcripts and/or genomes. A single APOBEC-induced C to U change in the infecting genomic (+) strand ssRNA would amplify in all viral progeny (C to U mutations). An ADAR-induced A to I (inosine) change in the (+) strand dsRNA would not reproduce in genomes of viral progeny. In contrast, an ADAR-induced A to I change in the (-) strand dsRNA would be copied into multiple (+) strand RNA transcripts and thus be amplified in the viral progeny as U to C mutations in genomic (+) strand ssRNA. (B) Viruses with double-stranded (ds) RNA genomes. Multiple (+) ssRNA transcripts and/or genome precursors are generated by RdRp. Each (+) ssRNAs precursor is then used to generate a dsRNA genome. Only ADAR-induced A to I mutations in (−) strand are amplified into multiple dsRNA genomes via copies of (+) strands. Since there are multiple (+) strand intermediates, there is a chance of detectable level of C to U APOBEC-induced deamination in a fraction of (+) strands. (C) Viruses with negative (−) ssRNA genomes. Several (+) ssRNA transcripts and/or precursors of (−) ssRNA genomes are generated by RdRp that are then used to generate multiple (−) ssRNA genomes. (−) ssRNA genomes of infecting particles as well (+) ssRNA precursors can serve as a substrate for APOBEC mutagenesis. The change (C to U or G to A) recovered by sequencing progeny genomes would be defined by the strand which is deaminated by APOBEC. Multiple C to U mutant molecules will arise from a single deamination in the infecting (−) ssRNA genome. Smaller number of G to A changes would result from each deamination event in a (+) strand precursor, but since there may be multiple precursor copies (shown in the multiple columns), a number of these changes may be comparable with C to U changes.