Figure 5.
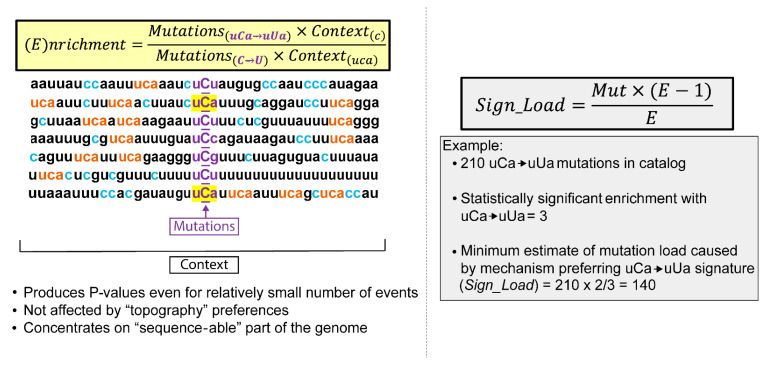
Trinucleotide motif-centered RNA mutational signature analysis. Shown is an example of an analysis for calculating the enrichment (E) and signature-associated mutation load (Sign Load) of uCa→uUa signature motifs in ssRNA, which are the two main outputs of trinucleotide motif-centered mutational signature analysis. Reverse complements are not included. Counted are all C→U mutations as well as all trinucleotide motif uCa→uUa mutations (5′ and 3′ flanking nucleotides shown in small letters; mutated C shown in capital letters). Also counted are all cytosines (c), represented in blue, and all motif-conforming trinucleotides (uca), represented in orange, in 41 nucleotide contexts centered around mutated cytosines. (E) values show the fold-difference between actual fraction of uCa→uUa mutations among C→U mutations in all trinucleotide motifs and the fraction of motif conforming trinucleotides (uca) among all cytosines (c) in the immediate vicinity of mutated cytosines. Counts used for enrichment calculation can be also used for calculating p-values in order to identify trinucleotide mutational motifs with statistically significant enrichment. Statistically significant enrichment values can be used for minimum estimates of a (Sign Load).