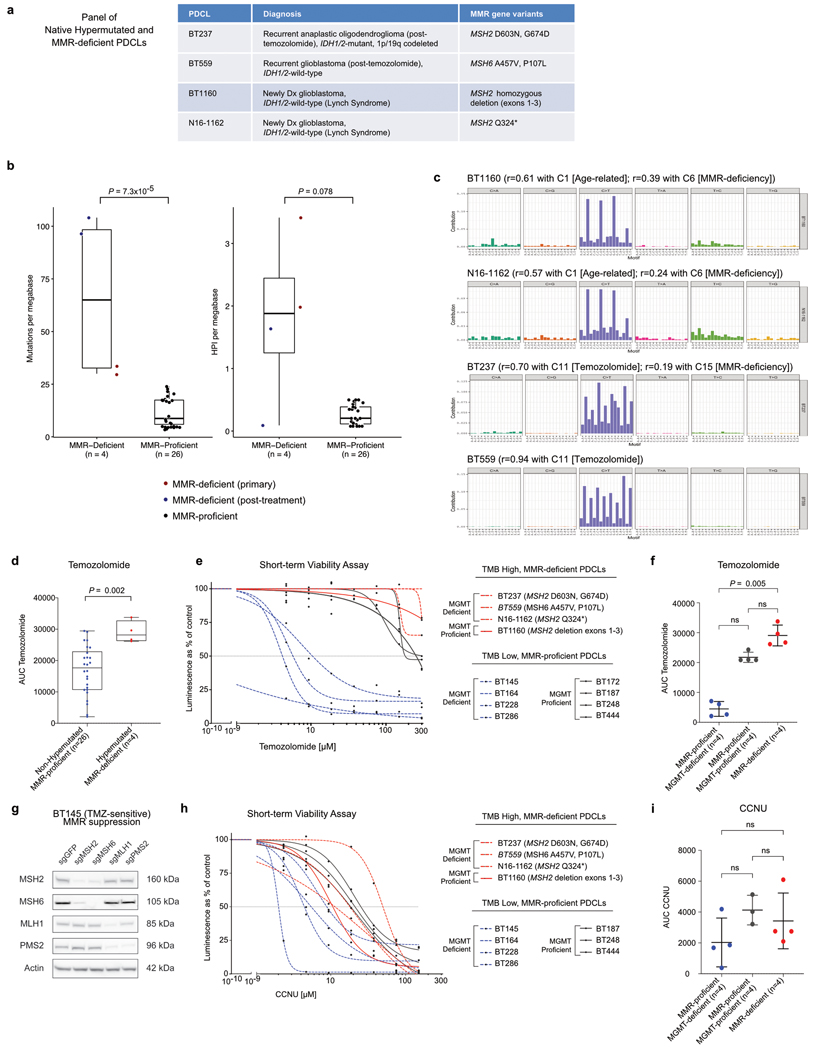
Extended Data Figure 8. Characterization of High-grade Glioma PDCLs and their Sensitivity to Temozolomide and CCNU.
a, Clinico-molecular characteristics of four native newly diagnosed or recurrent glioma PDCL models harbouring hypermutation and MMR deficiency. b, Thirty glioma PDCLs, including four PDCLs derived from patients with de novo (BT1160, N16-1162, both established from patients with Lynch syndrome) or post-treatment (BT237, BT559) MMR deficiency were molecularly characterized using whole-exome sequencing. The panels show the tumour mutational burden (left) and homopolymer indel burden (right) in each model. Boxes, quartiles; centre lines, median ratio for each group; whiskers, absolute range. Two-sided Wilcoxon rank-sum test. c, Mutational signature analysis was performed in the PDCL models of constitutional and post-treatment MMR deficiency using the R package DeconstructSigs to estimate the contributions of mutational signatures using a regression model (Rosenthal et al. 52). For each PDCL, the contribution of the main COSMIC mutational signatures identified is expressed as decimal. d, Boxplots of temozolomide AUC in non-hypermutated versus hypermutated PDCLs. Boxes, quartiles; centre lines, median ratio for each group; whiskers, absolute range. Two-sided Wilcoxon rank-sum test. e, f, A panel of 12 glioma PDCL models representing the different MGMT and MMR classes was selected and assessed for sensitivity to temozolomide in a short-term viability assay (e; dots represent means). The temozolomide AUC was compared between the three groups using a Kruskal–Wallis test and Dunn’s multiple comparison test (f; mean ± s.d.). g, Western blot of the glioblastoma patient-derived cell line (BT145) in which the genes MSH2, MSH6, MLH1 or PMS2 have been knocked-out using the CRISPR–Cas9 system. h, i, A panel of 11 glioma PDCL models representing the different MGMT and MMR classes was selected and assessed for sensitivity to CCNU in a short-term viability assay (h; dots represent means). No CCNU data was available for the model BT172. The CCNU AUC was compared between the three groups using a Kruskal–Wallis test and Dunn’s multiple comparison test (i; mean ± s.d.).