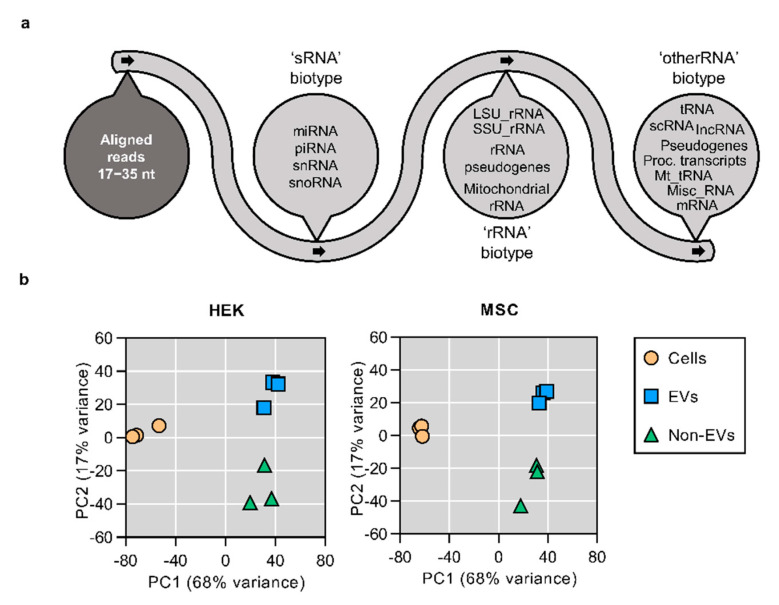
Figure 2.
Sequencing workflow and Principal Component Analysis of all uniquely annotated reads. (a) RNA was extracted from EV and Non-EV fractions and subjected to sequencing. RNA sequences with a read length of 17–35 nt were aligned to the human genome and annotated in a stepwise manner first to the ‘sRNA’ biotype (miRNA, piRNA, snRNA, snoRNA), then to the ‘rRNA’ biotype (LSU_rRNA, SSU_rRNA, rRNA pseudogenes, mitochondrial rRNA) and finally to the ’otherRNA’ biotype (tRNA, scRNA, lncRNA, pseudogenes, processed transcripts, mt_tRNA, misc_RNA, mRNA). (b) Principal Component Analysis of all uniquely annotated reads of both HEK (left panel) and MSC (right panel) samples showed clear cluster separation of the cell, EV and Non-EV samples. Looking at the principal component 1 (PC1, explaining 68% of variance), the secretory fractions of both samples are more similar to each other than to parent cells.