Abstract
Extensive pig systems are gaining importance as quality production systems and as the standard for sustainable rural development and animal welfare. However, the effects of natural foods on Salmonella epidemiology remain unknown. Herein, we assessed the presence of Salmonella and the composition of the gut microbiota in pigs from both Salmonella-free and high Salmonella prevalence farms. In addition, risk factors associated with the presence of Salmonella were investigated. The pathogen was found in 32.2% of animals and 83.3% of farms, showing large differences in prevalence between farms. Most isolates were serovars Typhimurium monophasic (79.3%) and Bovismorbificans (10.3%), and exhibited a multi-drug resistance profile (58.6%). Risk factor analysis identified feed composition, type/variety of vegetation available, and silos’ cleaning/disinfection as the main factors associated with Salmonella prevalence. Clear differences in the intestinal microbiota were found between Salmonella-positive and Salmonella-negative populations, showing the former with increasing Proteobacteria and decreasing Bacteroides populations. Butyrate and propionate producers including Clostridium, Turicibacter, Bacteroidaceae_uc, and Lactobacillus were more abundant in the Salmonella-negative group, whereas acetate producers like Sporobacter, Escherichia or Enterobacter were more abundant in the Salmonella-positive group. Overall, our results suggest that the presence of Salmonella in free-range pigs is directly related to the natural vegetation accessible, determining the composition of the intestinal microbiota.
Keywords: Salmonella, microbiota, free-range pigs, risk factors, antimicrobial resistance
1. Introduction
Salmonella enterica is one of the most common causes of food-borne zoonotic diseases worldwide. A total of 87,923 cases were reported in the European Union (EU) during 2019, making it the second most important zoonosis after campylobacteriosis [1]. Eggs and poultry products have been considered the most important source of human infections, responsible for 43.8% of cases [2], but the implementation of Salmonella control programs for fowl have resulted in decreasing trends in the occurrence of Salmonella in eggs in the EU. Consequently, a decrease in human salmonellosis has been reported from 2008 to 2019 [3]. Currently, pigs are the most important source of infection in humans, which has become a serious public health problem that requires special monitoring and surveillance. To preserve consumers’ health, the EU is discussing the establishment of restrictions in the international trade of pigs and pig products with countries that do not meet the objective of reducing the prevalence of Salmonella. These restrictions may have a major economic impact in our country because the pig sector is a key pillar of livestock resources in Spain, which is the fourth largest producer worldwide after the US, China and Germany [4]. Carrier animals are a serious food safety issue because they potentially shed the pathogen in feces, thereby contaminating other animals, slaughterhouses and meat products during processing. Additionally, the control of Salmonella in the food chain is impeded by the existence of over 2500 serovars, its broad host range and ubiquitous nature and its ability to sub-clinically colonize animals intended for human consumption.
In the recent years, consumers have become more aware of the way food is produced [5]. The increasing intensification of farming has been perceived negatively, while “animal-friendly” production such as free range is considered positive. Thus, extensive production has several features perceived by consumers as an improvement when compared with intensive management: (i) The animals mainly consume pasturage, reducing the need for industrial feed; (ii) It tends to raise autochthonous breeds that are well-adapted to the conditions of the land and extensive management; (iii) The production system is a sustainable model involving familiar farms; (iv) It provides the animals with the possibility of showing their natural behaviour; (v) It allows for low antibiotic usage; (vi) Due to natural food intake, the meat has a higher proportion of saturated/unsaturated fat, being healthier than intensive breeding; and (vii) The number of animals has to be kept in proportion with the available land due to the need for pastures, as well as slurry assimilation by the land and the excrements used as fertilizers. Extensive production promotes environmental sustainability and the development of rural areas.
The endogenous microbiota is known to provide important benefits to its host [6], and there is growing evidence suggesting that interactions between members of the gut microbiota of the host contribute to the health and well-being of these animals [7]. Recent studies have shown that Salmonella enterica is a pathogen capable of causing alterations to the composition of the intestinal microbiome, reducing the diversity and abundance of species regarded as beneficial such as lactic acid bacteria [8,9]. However, most of these studies are based in experiments where pigs have been challenged with the pathogen, and very few studies have focused on natural exposure of pigs to the pathogen; this is especially true for free range pigs. Thus, the main goal of this study is to widen the knowledge available on the microbiota of pigs raised in extensive systems in relation to the presence/absence of Salmonella, determine the prevalence of Salmonella spp. in this population, and decipher the risk factors associated with Salmonella spp. prevalence in pigs raised in outdoor systems.
2. Materials and Methods
2.1. Study Design
In order to study the presence of Salmonella in the intestinal contents (IC) of free-range fattening pigs (Duroc), 12 out of the 32 farms located in the Basque Country and north of Navarre were selected. They were visited between January and May 2015. The peculiarity of these farms was that animals were fed with cereals and food supplements ad libitum that were obtained directly from the field (pasture, acorns, chestnuts, or beechnuts).
All the animals of this study were transported to a slaughterhouse located less than 100 km (less than 2 h) from the farms, and they were slaughtered within 15–18 h of fasting. The intestinal contents (IC) of the cecum were collected from 15 animals/farm (n = 180 samples), randomly selected in the slaughterhouse line. The selection of cecum samples was based on the disruption to the microbiota in this anatomical location when challenged with Salmonella Typhimurium, as described by Borewicz et al. [9]. The whole intestinal package was removed from the selected carcasses at the evisceration point of the slaughter line, and 30 g of IC were collected aseptically in one-use sterile bottles, transported at 4 °C to the laboratory and immediately processed for Salmonella isolation (25 g). One gram per sample was frozen at −80 °C for microbiota analysis.
Ethics committee approval was not required because the work was performed with the entrails obtained in the conventional slaughtering line. Animal handling and slaughtering procedures were performed by the slaughterhouse personnel according to the current national legislation (Law 32/2007) for animal care regarding holding, transportation, testing and slaughtering.
2.2. Isolation of Salmonella
Salmonella isolation procedures were performed according to ISO 6579:2002/Amd 1:2007 (ISO) rules [10], as previously described [11,12,13]. All culture media and reagents used were purchased from Laboratorios Conda S.A., Spain. The IC samples (25 g) were homogenized in sterile filter bags (Stomacher® 80, Seward Medical) with 225 mL of Buffer Peptone Water for pre-enrichment. After the different ISO steps, one colony of Salmonella was sent for serotyping at the National Reference Laboratory for Animal Salmonellosis, Central Veterinary Laboratory (LCV, Madrid, Spain), following the Kauffmann–White Scheme [14].
Next, all samples were analyzed using a specific Salmonella PCR-invA [15] with the DNA extracted by boiling (95 °C, 10 min) from a 10 µL loop of MRSV that was mixed with 500 µL ultra-pure sterile water and showed the halo typical of Salmonella. Thereafter, samples showing positive results by PCR-invA and negative by ISO were re-analyzed microbiologically for Salmonella isolation.
All isolates were serotyped at the reference LCV (see above) and stored in the IdAB-CSIC collection by freezing in sterile, 10% skim milk (BD Difco™ Skim Milk, Sparks, MD, USA) supplemented with 3% lactose (Merck, Billerica, MA, USA).
The agreement between microbiological ISO and PCR-invA diagnosis was calculated using the kappa (κ) index and was interpreted according to the criteria of Landis and Koch [16].
2.3. Antimicrobial Susceptibility Testing
Confirmed Salmonella isolates were tested by the Kirby–Bauer disk diffusion method, using the antimicrobials (BD Diagnostics) and concentrations recommended by the current EU legislation for the harmonized monitoring of the antimicrobial resistance of Salmonella [17], namely, ampicillin (A), chloramphenicol (C), streptomycin and gentamicin (S), sulfisoxazole and trimethoprim–sulfamethoxazole (Su), tetracycline (T), nalidixic acid (Nx), enrofloxacin and cefotaxime (C3G). Antimicrobial susceptibility was determined by measuring the inhibition halo generated after incubation (37 °C, 24 h). Strains Escherichia coli ATCC 25,922, Salmonella Typhimurium DT104 and ATCC 14028 were used as controls. Isolates were classified as resistant or susceptible according to Clinical and Laboratory Standard Institute (CLSI) recommendations [18]; those exhibiting resistance to drugs of three or more antimicrobial families were considered multi-drug resistant (MDR).
2.4. Epidemiological Study and Risk Factors
Risk factors associated with Salmonella presence in pigs’ IC were studied by performing an epidemiological survey, with 73 variables divided into five sections. The variables analyzed were related to the geographical location of farms, climate, existing natural vegetation, type of feed, administration of antibiotics and general management. In addition, another survey was designed to analyze productive parameters such as weight at slaughter, slaughterhouse input weight, channel weight, average daily gain and feed conversion rate. These surveys were completed in collaboration with the veterinarians of the association and the personnel of the slaughterhouse.
All statistical analyses were carried out using SAS® V.9.1.3 (SAS institute Inc., Cary, NC, USA), setting p ≤ 0.05 as the significance level. For all analyses, the farm was considered the experimental unit. After conducting the surveys, the variables were coded, databases were elaborated in Excel® and the relevant statistical comparisons were carried out. First, Shapiro–Wilk and Levene tests were applied to assess if the continuous variables followed a normal distribution. The variables with a non-normal distribution were compared by non-parametric tests (Wilcoxon test). In the case of categorical variables, a Chi-square test (χ2) or Fisher’s exact test was used to assess the possible association among the variables. The risk factors associated with Salmonella prevalence at the farm level were determined by considering the percentage of infected animals on the farm as a continuous variable (n = 180) or as a categorical variable, considering a 20% prevalence cut-off (i.e., low or high prevalence on farms showing Salmonella in less than or more than 20% of pigs, respectively). Moreover, the variables detected as relevant (p < 0.2) in the univariable analysis were assessed for associations, applying X2 or an exact Fisher’s test in the case of categorical variables and collinearity analysis in the case of continuous variables. Finally, only variables that showed no association were included in a multivariable analysis. Thus, Salmonella prevalence (continuous variable) on a farm was analyzed by multivariable regression analysis, whereas low or high prevalence (categoric variable) was analyzed by a logistic, multivariable analysis.
2.5. Microbiota Analyses of Cecum Content
A total of 35 IC samples were selected to analyze the intestinal microbiota composition in animals from farms with very high or null prevalence of Salmonella spp. Thus, 10 and 5 pigs positive for Salmonella from two different farms and 20 pigs from two negative farms (10 animals/farm) were selected for microbiota analyses. The selection of farms was based on geographical location, type of food and presence/absence of Salmonella on the premises.
2.6. Intestinal Content DNA Extraction, PCR Amplification and Microbiota Analyses
Bacterial DNA was extracted from 0.2 mg of each sample using the PowerSoil™ DNA isolation kit (MO BIO) under manufacturer’s conditions. DNA samples were treated with 100 μL of the elution buffer and stored at −20 °C until further processing. The V1–V2 regions of 16S rRNA genes were amplified with primer pairs F27 (5′-AGAGTTTGATCCTGGCTCAG-3′) and R338 (5′-TGCTGCCTCCCGTAGGAGT-3′). Both primers included sequencing adaptors at the 5′ end, and forward primers were tagged with different barcodes. The PCR mixture (50 μL) contained 2 μL DNA template (~5 ng), 5 μL of 10x AccuPrime™ PCR Buffer II, 0.2 μM of each primer and 1 U of AccuPrime™ Taq DNA Polymerase High Fidelity (Invitrogen, Life Technologies, Carlsbad, CA, USA). The PCR thermal profile was 2 min at 94 °C followed by 30 cycles of 1 min at 94 °C, 1 min at 55 °C, 1 min at 72 °C and a final extension of 7 min at 72 °C. To check the absence of reagents contamination, each PCR included a negative control. For each amplicon, both concentration and quality were determined using Agilent Bioanalyzer 2100. Samples were sequenced on an Ion Torrent Personal Genome Machine (PGM) with the Ion 318 Chip Kit v2 (Life Technologies) and the Ion PGMTM Sequencing 400 Kit (Life Technologies) under manufacturer’s conditions. The raw sequences were deposited in NCBI under the Bioproject accession number PRJNA723169.
2.7. Quality Control, Operational Taxonomic Unit (OTUs), Diversity and Composition Analyses
Raw reads were imported into the EzBioCloud 16S-based MTP (microbiome taxonomic profiling) pipeline [19], which was used to analyze the whole dataset. Low quality sequences were filtered out using the following criteria: (i) read length less than 100 bp or more than 2000 bp; (ii) averaged Q value < 25; (iii) not predicted as a 16S rRNA gene by the hidden Markov model-based search; or (iv) detected as a singleton when sequences with at least 97% similarity did not match any of the reference sequences from the 16S database using the UCLUST program. Quimeric sequences were removed using the UCHIME program [20].
Taxonomic assignment was performed using the VSEARCH program [21] to detect and calculate the sequence similarities of the queried single-end reads against the EzBioCloud 16S database (version PKSSU4.0); 97% 16S similarity was used as the cut-off for species-level identification. Single-end reads from each sample were clustered into OTUs using the open-reference method [19].
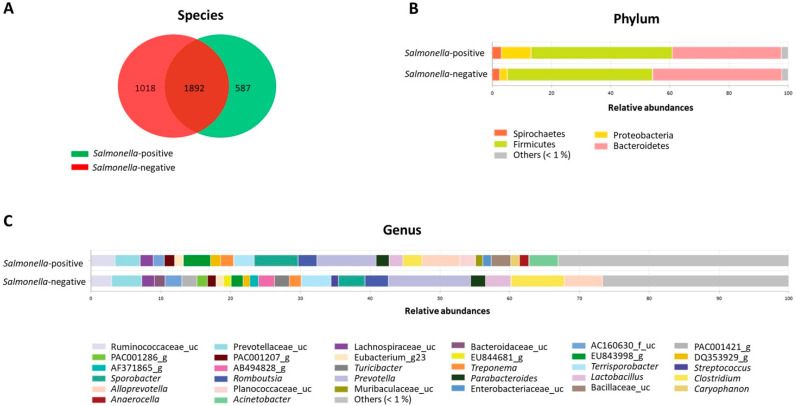
Alpha diversity was estimated by the OTU richness of each sample, measured by abundance-based coverage estimation (ACE) and Chao1 methods. Diversity was estimated by the Shannon, Simpson and Phylogenetic Diversity indices. Statistical significance of the alpha diversity values between Salmonella -positive and -negative groups was assessed using the Wilcoxon rank sum test. A p value < 0.05 was considered statistically significant. A Venn diagram plot was drawn using Venn diagram software (available online: http://bioinformatics.psb.ugent.be/webtools/Venn/, accessed on 17 June 2021) to compare the species among Salmonella -positive and -negative groups.
Beta diversity was assessed with UniFrac distances based on taxonomic abundance profiles. Statistical significance of the beta diversity clustering by Salmonella -positive and -negative groups was assessed using permutational multivariate analysis of variance (PERMANOVA) (p < 0.05). UniFrac distance matrices were used to perform principal coordinate analysis (PCoA) with the Qiime2 tool [22] to compare the microbial communities among the groups. Additionally, differences in beta diversity were evaluated among farms.
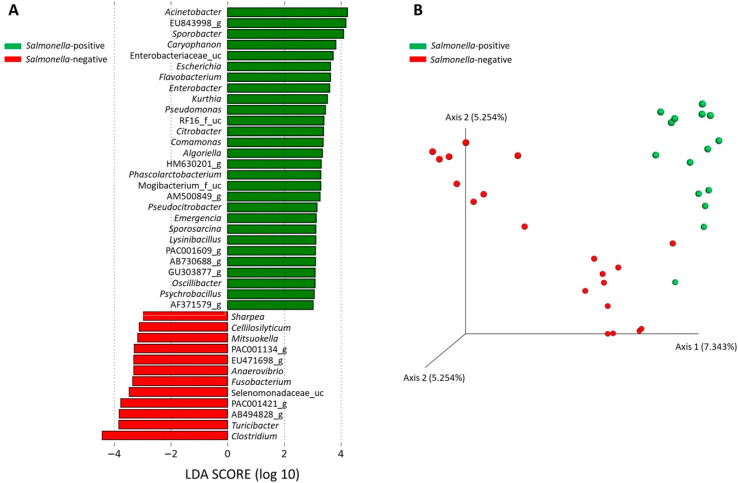
Linear discriminant analysis (LDA) effect size (LEfSe) [23] was used to compare Salmonella -positive and -negative groups and to identify statistically significant differences in taxa abundances between groups. Taxonomic levels with LDA score > 2 and p value < 0.05 were statistically significant.
3. Results
3.1. Prevalence of Salmonella in the Extensive System
The presence of Salmonella in the IC from free-range pigs was studied using both the ISO as and PCR-invA methods and confirmed by the isolation and typing of the pathogen. The standard ISO allowed us to detect Salmonella in 47 out of 180 (26.1%) IC samples, whereas PCR-invA detected the pathogen in the same samples, as well as in an additional 11 (Table S1). The concordance test between both diagnostic techniques was almost perfect (k = 0.851) due to the high number of negative samples in both techniques (Table S1).
Considering both diagnostic methods, Salmonella was detected in the feces of 32.2% (58/180) of free-range pigs, allocated among 10 out of the 12 (83.3%) farms sampled (Table 1). Also, the mean prevalence within Salmonella-positive farms (i.e., herds containing at least one positive pig) was 38.7%, showing that for most of the farms (66.7%), more than 20% of the pigs shed Salmonella in their feces (Table 1 and Table S2).
Table 1.
Detection of Salmonella by the ISO 6579:2002/Amd 2007 and/or molecular methods in the intestinal content of free-range pigs of North Spain.
| Salmonella spp. Isolation | Intestinal Content |
|---|---|
| No. (%; CI95) 1 positive pigs 2/total pigs | 58/180 (32.2%; 25.8–39.3) |
| No. (%; CI95) 1 positive farms 2/total farms | 10/12 (83.3%; 55.2–95.3) |
| No. (%; CI95) 1 positive pigs 2/pigs in positive farms | 58/150 (38.7; 31.2–46.6) |
| No. (%) farms above 20% prevalence/total farms | 8/12 (66.7%; 39.1–86.2) |
1 mean % and 95% of confidence interval; 2 positive pigs or farms were those where at least 1 Salmonella positive sample was detected.
3.2. Serotyping and Antimicrobial Susceptibility
As shown in Table 2, the 58 Salmonella isolates belonged to 6 different serovars, being S. Typhimurium monophasic variant (4,[5],12:i:-) the most frequently found (46/58; 79.3% pigs) followed by far by S. Bovismorbificans (6/58; 10.3% pigs). Interestingly, the monophasic variant was found in 8/10 positive farms, being the only serovar circulating in 6 of these farms with the highest number of infected animals, varying between 46.7% and 86.7% (Table 2). Regarding antimicrobial susceptibility, 89.7% (52/58) of isolates exhibited resistance to at least one of the antimicrobials tested, being streptomycin (51/58; 87.9%), sulfisoxazole (44/58; 75.7%), ampicillin (31/58; 53.4%) and tetracycline (24/58; 41.4%), the drugs with the highest proportion of resistant isolates. Additionally, 58.6% (34/58) of them showed different multi-drug resistant profiles. Thirteen out of the 46 (28.2%) monophasic variants exhibited the ASSuT profile (Table 2), although other profiles were also detected such as the penta-resistant phenotype (ACSSuT).
Table 2.
Prevalence of Salmonella in the intestinal content of free-range pigs. Data are presented by farms, serovars and antimicrobial resistance (AR) profiles of the isolates found.
| Farm Code | No. (%) Samples Positive/Total | Serovars (No. of Strains) | AR Profiles 1 (No. of Strains) |
|---|---|---|---|
| 1 | 13/15 (86.7%) | 4,5,12:i:- (13) | SSu (5); ASSuT (4); ASSu (2); ACSSuT (2) |
| 2 | 8/15 (53.3) | 4,5,12:i:- (8) | ACS (3); CS (2); ACSSu (1); AS (1); ACSSuT (1) |
| 3 | 7/15 (46.7%) | 4,5,12:i:- (7) | SSu (4); ASSuT (3) |
| 4 | 7/15 (46.7%) | 4,5,12:i:- (7) | ASSu (4); ASSuT (2); SSu (1) |
| 5 | 6/15 (40%) | 4,5,12:i:- (5); diarizonae (1) |
ASSuT (3); SSu (1); A (1) Susceptible (1) |
| 6 | 5/15 (33.3%) | Bovismorbificans (4); Altona (1) |
ACSSuT (2); CSSuT (2); S (1) |
| 7 | 4/15 (26.7%) | Bovismorbificans (2); Meleagridis (2); |
CSSuT (2); Susceptible (2) |
| 8 | 4/15 (26.7%) | 4,5,12:i:- (2); Amsterdam (1); Altona (1) |
ASSuT (2); Susceptible (2) |
| 9 | 3/15 (20%) | 4,5,12:i:- (3) | ASSu (2); Susceptible (1) |
| 10 | 1/15 (6.7%) | 4,5,12:i:- (1) | ASSuT (1) |
| 11 | 0/15 (0%) | N.A. | N.A. |
| 12 | 0/15 (0%) | N.A. | N.A. |
| Total | 58/180 (32.2%) | 6 serovars (58) | 11 AR profiles |
1 A: ampicillin; C: chloramphenicol; S: streptomycin; Su: sulfisoxazole and/or trimetoprim–sulfometoxazole; T: tetracycline; N.A.: not applicable.
3.3. Risk Factors Associated with the Presence of Salmonella in IC Samples
Once the questionnaires were filled out at the farms and slaughterhouse, variables were coded and data was debugged. Questions with no answers or answers without variability were removed from the statistical analysis. No significant differences were detected among the five productive parameters (mean ± SD) analyzed: piglet weight at the beginning of the fattening phase (29.3 kg ± 3.9), input weight at the slaughterhouse (154.3 kg ± 8.7), carcass weight (118 kg ± 6.6), daily gain mean (773.6 ± 54.3) and feed conversion rate (3.76 ± 0.238). As a result, a total of 52 variables were analyzed, and only those risk factors that showed p ≤ 0.2 in the univariable analysis were included in the multivariable analysis. Thus, four variables were associated with a high risk of detecting Salmonella in IC, considered categorical (Table 3); (i) large farms (>100 pigs); (ii) farms that used Feed A (with soybeans and beet pulp) versus Feeds B and C (beans and rapeseed meal, respectively); (iii) farms with exclusively grass-based vegetation versus those with chestnuts, acorns and beeches available; and (iv) farms with silos that were neither cleaned nor disinfected. These last three variables were associated with each other (p < 0.05), and the feed and extra vegetation available to the animals were selected to be included in the multivariable analysis. Details about these variables by farm are included as supplementary materials (Table S2). Thereafter, the multivariable analysis was performed by introducing into the model the “feed composition” (Feed A vs. B/C) and the “holding size” (>100 vs. ≤100 animals/farm) as independent variables. The logistic model indicated that feed A (soybeans and beet pulp) was a significant risk factor (p = 0.008) associated with the presence of Salmonella (Table 3).
Table 3.
Risk factors associated with salmonellosis in free-range pigs. The prevalence of Salmonella on the farm was considered to be a continuous variable or as a categorical variable considering the cut-off of 20% prevalence (low or high prevalence on farms with presence of Salmonella spp. in less than or more than 20% of pigs, respectively). The analysis of the prevalence as a continuous variable or as a categorical was carried out by a multivariable regression or logistic analysis, respectively.
| Risk Factors | Statistical Analysis (p Values) | |||
|---|---|---|---|---|
| Univariable | Multivariable | |||
| Prevalence | Low/High | Prevalence | Low/High | |
| (i) Large farms (>100 animals) | 0.02 | 0.06 | 0.11 | 0.19 |
| (ii) Feed with A brand | 0.13 | 0.02 | 0.29 | 0.008 |
| (iii) Extra feed with grass | 0.38 | 0.02 | N.A. | N.A. |
| (iv) Neither cleaning nor disinfection of silos | 0.04 | 0.02 | N.A. | N.A. |
N.A.: not applicable because these variables were associated to risk factor “ii”.
3.4. Microbiota Analysis in Two Pig Populations
As shown in Table S2, a total of 35 fecal samples were selected to study their microbiota composition, distributed between two groups of 15 Salmonella-positive pigs from two different farms (10 and 5 pigs/farm) and 20 Salmonella-negative pigs from two negative farms (10 animals/farm). A mean of 50,621 reads per sample were analyzed. A total of 21 phylum, 55 classes, 117 orders, 265 families, 987 genera and 3497 species were identified at a 97% sequence similarity level. A mean number of 2479 species were detected in the Salmonella-positive samples compared with 2910 detected in the Salmonella-negative samples. A total of 1897 OTUs were in both groups; however, 587 and 1018 were unique for Salmonella-positive and -negative samples, respectively (Figure 1A). No significant differences were observed in species richness between both groups using either the Shannon or Simpson tests (Table 4), but a significantly higher phylogenetic diversity index was obtained in the Salmonella-positive group (p = 0.044).
Figure 1.
Microbial community comparisons between Salmonella-positive and -negative groups. (A) Venn diagram showing unique and shared species. (B) Relative abundance of bacterial phyla. (C) Relative abundance of bacterial genera.
Table 4.
Operational taxonomic units (OTUs)’ richness and alpha diversity indexes estimated in the gut microbiota of Salmonella-positive and Salmonella-negative pigs.
| Parameter | OTU Richness and Alpha Diversity Indexes Median and (IQR) a Values |
||
|---|---|---|---|
| Salmonella-Positive Pigs | Salmonella-Negative Pigs | p Values | |
| OTU number | 3132 (583) | 3047 (714.73) | 0.271 |
| Chao1 | 3156.06 (581.84) | 3085.74 (725.19) | 0.301 |
| ACE b | 3227.75 (580.90) | 3166.53 (725.19) | 0.301 |
| Shannon | 6.44 (0.64) | 6.04 (0.80) | 0.072 |
| Simpson | 0.01 (0) | 0.01 (0.01) | 0.062 |
| Phylogenetic diversity | 1366 (80) | 1288 (119.50) | 0.044 c |
a IQR: interquartile range; b ACE: abundance-based coverage estimation; c p < 0.05 between both pig groups by Wilcoxon rank sum test.
The analysis of microbial community composition showed that the predominant phyla in the Salmonella-positive and -negative groups were Firmicutes (47.79% and 49.01%, respectively) and Bacteroidetes (36.81% and 43.67%, respectively). The relative abundance of Firmicutes was similar in both groups, whereas the abundance of Bacteroidetes was significantly lower and the abundance of Proteobacteria was higher in the Salmonella-positive group (Figure 1B). The presence of 20 bacterial families was significantly differentiated among the two groups. The families more represented in the Salmonella-positive group were Moraxellaceae, Planococcaceae, Ruminococcaceae, Enterobacteriaceae and RF16_f in contrast to Clostridiaceae, AC160630_f and Selenomonadaceae in the Salmonella-negative group.
At the genera level (Figure 1C), Prevotella was the most abundant in both groups, with a relative abundance of 8.43% and 11.58% in the Salmonella-positive and -negative groups, respectively. Sporobacter (6.23%) was the second most abundant genus in the Salmonella-positive group and Clostridium (7.5%) in the Salmonella-negative. LEfSe analysis (LDA score > 2.0, p < 0.05) showed that taxa abundance was significantly different for 28 and 12 genera in Salmonella-positive and Salmonella-negative pigs, respectively (Figure 2A). Acinetobacter, EU843998_g, Sporobacter, Caryophanon, Enterobacteriaceae_uc, Escherichia, Flavobacterium and Enterobacter were represented in the microbiota of the Salmonella-positive group, whereas Clostridium, Turicibacter, AB494828_g and PAC001421_g, among others, were significantly represented in the Salmonella-negative group.
Figure 2.
Differential bacterial taxa analysis in Salmonella-positive (green) and Salmonella-negative (red) groups. (A) Distinctive genera of each group obtained by LEfSe analysis (LDA score > 2.0; alpha value < 0.05). (B) Principal coordinate analysis (PCoA) graph based on UniFrac distances.
Finally, UniFrac-based PERMANOVA analysis indicated significant differences (p = 0.006) between Salmonella-positive and -negative groups. A three-dimensional PCoA graph (Figure 2B) based on UniFrac distances (ANOSIM value R = 0.05, p < 0.001) showed well-defined and different microbiota compositions for each group. Significant differences were also observed in beta diversity among farms classified as Salmonella-negative (p = 0.013); while Clostridiales, Erysipelotrichales and Selenomonadales were highly represented in farm 12, the Spirochaetales order was more abundant in farm 10.
4. Discussion
The prevalence of Salmonella in pigs reared in outdoor systems has been poorly studied. The actual re-emergence of this type of production makes it necessary to study the prevalence of infection and the risk factors associated with the occurrence of Salmonella. Furthermore, studies on the microbiota composition in these animals have not been previously reported; as well as, variations of microbiota composition with regard to the presence/absence of Salmonella. In general, high Salmonella prevalence was observed in animals reared outdoors but with high differences between farms. Several studies have been carried out in outdoor systems; however, they reported different management practices including organic, non-organic or Iberian pigs, making these results hardly comparable to ours [24,25]. Contrarily to our findings, only a single antibiotic-free pig from the extensive system was positive for Salmonella spp. [24], and 5.3% of Iberian pigs belonging to 33% of the herds from South Spain were Salmonella-positive in their mesenteric lymph nodes (MLN) [25]. These differences in prevalence might be explained by the type and quantity of samples (i.e., 1 g of MLN vs. 25 g of feces used in our study) and could also be influenced by different weather and feeding conditions. Jensen and collaborators investigated the dynamics of Salmonella infection in organic pigs and its dispersion in the pasture, demonstrating that Salmonella persisted in the environment, contaminating grass then able to cause infections in pigs [26]. The longitudinal occurrence of Salmonella in outdoor systems in the UK provided evidence that movement to a new location in outdoor systems has an overall beneficial effect on Salmonella carriage [27], decreasing its prevalence in feces from 29.6% to 16.9%. This management practice could help to reduce Salmonella in outdoor herds. However, new land is not always available, and perhaps more frequent resting and rotation of the land used for paddock systems within a field site could be of help. Finally, in the UK and Denmark [27], pigs reared in outdoor systems showed similar Salmonella prevalence to those of our study (29.6%). The prevalence of salmonellosis reported herein was very similar to that reported by Spain [28] and Aragón [29], both with intensive indoor systems. To our surprise, extensive swine showed a much higher prevalence than the intensive fattening pig of Navarra (8.4% of the animals) [12].
Knowledge of the serovars and AR of circulating Salmonella strains is necessary both, for appropriate antibiotic treatments and for epidemiological follow-up, e.g., identification of the origin of contamination and control of the expansion of clones carrying mobile genetic elements with multi-drug resistant profiles. The emergence of the monophasic variant exhibiting multi-drug resistance, ASSuT [30], has been increasingly reported in the last few years [31] and is widely disseminated in the EU [32,33], including Spain (this study). The spread of multi-drug resistant Salmonella from farm to fork implies a major risk from a one-health perspective. For this reason, detecting risk factors associated with the presence of Salmonella in the primary sector is essential for designing mitigation strategies. Additionally, it is interesting that only the monophasic variant was recovered herein, while Salmonella Typhimurium (4,5:12:i:1,2) was the most prevalent serovar detected in fattening pigs and sows within intensive systems [11,12,29]. Furthermore, the most common serovars reported in Iberian pigs were Anatum and Typhimurium with resistance to streptomycin and tetracycline [25], differing from those described in our study.
The control of Salmonella requires determining those risk factors associated with a high prevalence in fattening pigs reared in outdoor systems. The number of farms analyzed herein (12 out of 32) were enough to determine the prevalence of Salmonella; however, the multivariable analysis was performed only with those variables that showed highly significant p values in the univariable model. Unfortunately, there were many variables included in the analysis, but not enough farms (low statistical potency). Moreover, several risk factors showed statistical association between them, impeding the evaluation of the relative weight of each variable in the model. Further work should be performed on those farms that have sufficient variability in associated parameters (feed compositions, different extra feed, silo cleaning) to validate the model.
Diet A (soybeans and beet pulp) was a risk factor that increased the prevalence of Salmonella. Perhaps there was defective or insufficient heat treatment of the granulate or cross-contamination during processing, storage and/or transportation that favored the growth of the pathogen. In addition, feed contamination may come from silos where inadequate cleaning and disinfection is carried out, especially if the silos are exposed to rodents, birds, insects and other free-living vectors [27,34,35]. On the other hand, the origin of the feed could also determine the quality of the ingredients (raw materials and/or additives) used in its formulation and physical structure. This aspect, associated to the nutritional quality of the surrounding vegetation, probably modulates the composition of the bacterial populations in the digestive tracts of pigs from extensive systems. Pigs fed with more nutritious products could develop a microbiota capable of preventing the presence of Salmonella in the intestine by creating a suitable microenvironment (digestive tract acidification) and/or the proliferation of other bacterial taxa that could promote competitive exclusion of the pathogen. To address this hypothesis, we analyzed the profile of the intestinal microbiota of a selected population with different types of food and the presence and absence of Salmonella.
The gut microbiota in pig cecum samples was largely composed of obligate anaerobic bacteria belonging to Firmicutes and Bacteroidetes, the main phyla detected herein, accounting for more than 88% of all bacteria in pigs, as observed by other authors [8,9]. These phyla degrade the nutrients present in the distal gut to a variety of metabolites producing short-chain fatty acids (SCFAs), with acetate, propionate and butyrate being the most abundant [36]. Interestingly, several studies have demonstrated the effects of propionate and butyrate production in the prevention of Salmonella colonization [37,38,39]. In fact, our results confirm higher abundance of Clostridium, Turicibacter, Bacteroidaceae_uc, and Lactobacillus in Salmonella-negative animals, all genera associated with the production of these two SCFAs [40]. Additionally, species of Lactobacillus have been proven to effectively inhibit the colonization of Salmonella by the secretion of metabolites with antimicrobial activity or by impairing the adhesion of Salmonella to the intestinal cells [41]. Contrarily, acetate production has been linked to Salmonella invasion [39] by inducing the expression of the genes located in pathogenicity island I (ISPI-1) required to penetrate the intestinal epithelial cells [42]. In this regard, we detected an enrichment of the genus Sporobacter, described as an acetate producer [43] in the Salmonella-positive group, together with Acinetobacter, which can utilize acetate as carbon source. In fact, an increase in the abundance of Proteobacteria, a phylum that has been associated with a microbial signature of dysbiosis in the gut and epithelial injury, was observed in Salmonella-positive animals [44,45]. Families of Moraxellaceae and Enterobacteriaceae belonging to the Gammaproteobacteria class were enriched in these animals, with the latter family containing a large number of potential pathobionts for humans and pigs including Salmonella, Escherichia and Enterobacter. Herein, V1–V2 amplification of the 16S rRNA region allowed us to detect differences in the abundance of Proteobacteria among Salmonella prevalence groups, despite the suggested limitation proposed by Johnson and collaborators [46]. Differences in beta diversity were also observed among pigs from farms negative for Salmonella, probably due to the dietary differences associated with a diet rich in carbohydrates and fiber in animals feeding on acorns as well as starch, provided mainly by the chestnuts.
The results obtained in this study indicated that diet conditioned the intestinal microbiota of pigs, a decisive factor in the presence/absence of Salmonella in the intestinal contents of these animals. Along these lines, future studies could contribute to elucidating the impact of different dietary ingredients, comparing those traditionally of good quality (barley, wheat and maize) with others containing large numbers of raw materials (soybeans, sorghum, cassava, pulp or beet molasses), which could potentially balance the porcine intestinal microbiota and thereby prevent the presence of Salmonella in the intestinal tract.
Acknowledgments
We thank the Basatxerri association slaughterhouse staff, Josu Garaialde and Rosa Larrañaga, as well as the veterinarians enrolled in this work: Imanol Rekondo, Isaías Bautista and Francisco Martín-Mendiburu. We also thank Cristina de Frutos from the National Reference Laboratory for Animal Salmonellosis, Central Veterinary Laboratory (LCV, Madrid, Spain) for the Salmonella serotyping.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/foods10061410/s1, Table S1: Contingency table with results of Salmonella identification by ISO 6579:2002/Amend. 1:2007 (ISO) and/or PCR-invA using MRSV as the DNA source (PCR); Table S2: Classification of farms by Salmonella prevalence in the intestinal content of free-range pigs and the risk factors associated by univariable analysis.
Author Contributions
Conceptualization: M.J.G.; Data curation: A.A.-G., I.M.-B.; Formal analysis: V.G., I.G., A.A.-G., I.M.-B., L.F.; Funding acquisition: M.J.G.; Investigation: V.G., I.G., M.J.G.; Methodology: V.G., M.J.G.; Project administration: M.J.G.; Resources: M.J.G.; Supervision: V.G., M.J.G.; Validation: V.G., L.F., L.M.-G., M.J.G.; Visualization: V.G., A.A.-G., I.M.-B., L.M.-G., M.J.G.; Writing—original draft: V.G., L.M.-G., M.J.G.; Writing—review & editing: all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Caja Navarra Foundation (project reference 70628).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated in this study is included in the article. The raw sequences were deposited in NCBI under the Bioproject accession number PRJNA723169. Further information on data and samples is available from the corresponding author on request.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.European Food Safety Authority. European Centre for Disease Prevention and Control The European Union One Health 2019 Zoonoses Report. EFSA J. 2021;19:e06406. doi: 10.2903/j.efsa.2021.6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pires S.M., de Knegt L., Hald T. Scientific/Technical Report Submitted to EFSA. National Food Institute, Technical University of Denmark; Søborg, Denmark: 2011. Estimation of the relative contribution of different food and animal sources to human Salmonella infections in the European Union. [Google Scholar]
- 3.EFSA-ECDC The European Union One Health 2018 Zoonoses Report. EFSA J. 2019;17:5926. doi: 10.2903/j.efsa.2019.5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MAPAMA Caracterización del Sector Porcino Español Año 2019. [(accessed on 17 June 2021)];2020 Available online: https://www.mapa.gob.es/es/ganaderia/temas/produccion-y-mercados-ganaderos/indicadoreseconomicoscarnedecerdo2019_tcm30-379728.pdf.
- 5.Thorslund C.A., Aaslyng M.D., Lassen J. Perceived importance and responsibility for market-driven pig welfare: Literature review. Meat Sci. 2017;125:37–45. doi: 10.1016/j.meatsci.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Backhed F., Ley R.E., Sonnenburg J.L., Peterson D.A., Gordon J.I. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 7.Isaacson R., Kim H.B. The intestinal microbiome of the pig. Anim. Health Res. Rev. 2012;13:100–109. doi: 10.1017/S1466252312000084. [DOI] [PubMed] [Google Scholar]
- 8.Bearson S.M., Allen H.K., Bearson B.L., Looft T., Brunelle B.W., Kich J.D., Tuggle C.K., Bayles D.O., Alt D., Levine U.Y., et al. Profiling the gastrointestinal microbiota in response to Salmonella: Low versus high Salmonella shedding in the natural porcine host. Infect. Genet. Evol. 2013;16:330–340. doi: 10.1016/j.meegid.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 9.Borewicz K.A., Kim H.B., Singer R.S., Gebhart C.J., Sreevatsan S., Johnson T., Isaacson R.E. Changes in the porcine intestinal microbiome in response to infection with Salmonella enterica and Lawsonia intracellularis. PLoS ONE. 2015;10:e0139106. doi: 10.1371/journal.pone.0139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.ISO . Animal Faeces and in Samples from the Primary Production Stage. ISO; Geneve, Switzerland: 2007. International Organisation for Standardisation. ISO 6579:2002/Amd. 1:2007. Microbiology of food and animal feeding stuffs. Horizontal method for the detection of Salmonella spp. Amendment 1 Annex D: Detection of Salmonella spp. [Google Scholar]
- 11.Garrido V., Sanchez S., San Roman B., Fraile L., Migura-Garcia L., Grillo M.J. Salmonella Infection in Mesenteric Lymph Nodes of Breeding Sows. Foodborne Pathog. Dis. 2020;17:411–417. doi: 10.1089/fpd.2019.2708. [DOI] [PubMed] [Google Scholar]
- 12.San Roman B., Garrido V., Sanchez S., Martinez-Ballesteros I., Garaizar J., Mainar-Jaime R.C., Migura-Garcia L., Grillo M.J. Relationship between Salmonella infection, shedding and serology in fattening pigs in low-moderate prevalence areas. Zoonoses Public Health. 2018;65:481–489. doi: 10.1111/zph.12453. [DOI] [PubMed] [Google Scholar]
- 13.Garrido V., Sanchez S., San Roman B., Zabalza-Barangua A., Diaz-Tendero Y., de Frutos C., Mainar-Jaime R.C., Grillo M.J. Simultaneous infections by different Salmonella strains in mesenteric lymph nodes of finishing pigs. BMC Vet. Res. 2014;10:59. doi: 10.1186/1746-6148-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimont P.A., Weill F.X. Antigenic formulae of the Salmonella Serovars. 9th ed. World Health Organization (WHO) Collaborating Centre for Reference and Research on Salmonella; Institut Pasteur; Paris, France: 2007. [Google Scholar]
- 15.Mainar-Jaime R.C., Atashparvar N., Chirino-Trejo M. Estimation of the diagnostic accuracy of the invA-gene-based PCR technique and a bacteriological culture for the detection of Salmonella spp. in caecal content from slaughtered pigs using Bayesian analysis. Zoonoses Public Health. 2008;55:112–118. doi: 10.1111/j.1863-2378.2007.01096.x. [DOI] [PubMed] [Google Scholar]
- 16.Landis J.R., Koch G.G. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 17.EU Commission Decision of 12 June 2007 on a Harmonised monitoring of antimicrobial resistance in Salmonella in poultry and pigs. 2007/407/EC. Official Journal of the European Union. Off. J. Eur. Union. 2007;50:L153. [Google Scholar]
- 18.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. Standard M100S. CLSI; Wayne, PA, USA: 2016. [Google Scholar]
- 19.Yoon S.H., Ha S.M., Kwon S., Lim J., Kim Y., Seo H., Chun J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rognes T., Flouri T., Nichols B., Quince C., Mahe F. VSEARCH: A versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thakur S., Tadesse D.A., Morrow M., Gebreyes W.A. Occurrence of multidrug resistant Salmonella in antimicrobial-free (ABF) swine production systems. Vet. Microbiol. 2007;125:362–367. doi: 10.1016/j.vetmic.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 25.Gomez-Laguna J., Hernandez M., Creus E., Echeita A., Otal J., Herrera-Leon S., Astorga R.J. Prevalence and antimicrobial susceptibility of Salmonella infections in free-range pigs. Vet. J. 2011;190:176–178. doi: 10.1016/j.tvjl.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 26.Jensen A.N., Dalsgaard A., Stockmarr A., Nielsen E.M., Baggesen D.L. Survival and transmission of Salmonella enterica serovar typhimurium in an outdoor organic pig farming environment. Appl. Environ. Microbiol. 2006;72:1833–1842. doi: 10.1128/AEM.72.3.1833-1842.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith R.P., Andres V., Dormer L., Gosling R., Oastler C., Davies R.H. Study of the impact on Salmonella of moving outdoor pigs to fresh land. Epidemiol. Infect. 2017;145:1983–1992. doi: 10.1017/S0950268817000930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.EFSA Report of the task force on zoonoses data collection on the analysis of the baseline survey on the prevalence of Salmonella in slaughter pigs. Part A. EFSA J. 2008;135:1–111. [Google Scholar]
- 29.Vico J.P., Rol I., Garrido V., San Román B., Grilló M.J., Mainar-Jaime R.C. Salmonellosis in finishing pigs in Spain: Prevalence, antimicrobial agent susceptibilities, and risk factor analysis. J. Food Prot. 2011;74:1070–1078. doi: 10.4315/0362-028X.JFP-10-515. [DOI] [PubMed] [Google Scholar]
- 30.Echeita M.A., Herrera S., Usera M.A. Atypical, fljB-negative Salmonella enterica subsp. enterica strain of serovar 4,5,12:i:- appears to be a monophasic variant of serovar Typhimurium. J. Clin. Microbiol. 2001;39:2981–2983. doi: 10.1128/JCM.39.8.2981-2983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biswas S., Li Y., Elbediwi M., Yue M. Emergence and Dissemination of mcr-Carrying Clinically Relevant Salmonella Typhimurium Monophasic Clone ST34. Microorganisms. 2019;7:298. doi: 10.3390/microorganisms7090298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.EFSA Scientific Report on the analysis of the baseline survey on the prevalence of Salmonella in slaughter pigs, in the EU, 2006-2007. Part B: Factors associated with Salmonella infection in lymph nodes, Salmonella surface contamination of carcasses, and the distribution of Salmonella serovars. EFSA J. 2008;206:1–111. [Google Scholar]
- 33.Petrovska L., Mather A.E., AbuOun M., Branchu P., Harris S.R., Connor T., Hopkins K.L., Underwood A., Lettini A.A., Page A., et al. Microevolution of Monophasic Salmonella Typhimurium during Epidemic, United Kingdom, 2005–2010. Emerg. Infect. Dis. 2016;22:617–624. doi: 10.3201/eid2204.150531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andres-Barranco S., Vico J.P., Garrido V., Samper S., Herrera-Leon S., de Frutos C., Mainar-Jaime R.C. Role of wild bird and rodents in the epidemiology of subclinical salmonellosis in finishing pigs. Foodborne Pathog. Dis. 2014;11:689–697. doi: 10.1089/fpd.2014.1755. [DOI] [PubMed] [Google Scholar]
- 35.Andres S., Vico J.P., Garrido V., Grillo M.J., Samper S., Gavin P., Herrera-Leon S., Mainar-Jaime R.C. Epidemiology of subclinical salmonellosis in wild birds from an area of high prevalence of pig salmonellosis: Phenotypic and genetic profiles of Salmonella isolates. Zoonoses Public Health. 2013;60:355–365. doi: 10.1111/j.1863-2378.2012.01542.x. [DOI] [PubMed] [Google Scholar]
- 36.Rivera-Chavez F., Baumler A.J. The Pyromaniac Inside You: Salmonella Metabolism in the Host Gut. Annu. Rev. Microbiol. 2015;69:31–48. doi: 10.1146/annurev-micro-091014-104108. [DOI] [PubMed] [Google Scholar]
- 37.Rivera-Chavez F., Zhang L.F., Faber F., Lopez C.A., Byndloss M.X., Olsan E.E., Xu G., Velazquez E.M., Lebrilla C.B., Winter S.E., et al. Depletion of Butyrate-producing clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe. 2016;19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chu B., Zhu Y., Su J., Xia B., Zou Y., Nie J., Zhang W., Wang J. Butyrate-mediated autophagy inhibition limits cytosolic Salmonella Infantis replication in the colon of pigs treated with a mixture of Lactobacillus and Bacillus. Vet. Res. 2020;51:99. doi: 10.1186/s13567-020-00823-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawhon S.D., Maurer R., Suyemoto M., Altier C. Intestinal short-chain fatty acids alter Salmonella Typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 2002;46:1451–1464. doi: 10.1046/j.1365-2958.2002.03268.x. [DOI] [PubMed] [Google Scholar]
- 40.Nishitsuji K., Xiao J., Nagatomo R., Umemoto H., Morimoto Y., Akatsu H., Inoue K., Tsuneyama K. Analysis of the gut microbiome and plasma short-chain fatty acid profiles in a spontaneous mouse model of metabolic syndrome. Sci. Rep. 2017;7:15876. doi: 10.1038/s41598-017-16189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hai D., Lu Z., Huang X., Lv F., Bie X. In Vitro Screening of Chicken-Derived Lactobacillus Strains that Effectively Inhibit Salmonella Colonization and Adhesion. Foods. 2021;10:569. doi: 10.3390/foods10030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones B.D., Ghori N., Falkow S. Salmonella Typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer’s patches. J. Exp. Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.GrechMora I., Fardeau M.L., Patel B.K.C., Ollivier B., Rimbault A., Prensier G., Garcia J.L., GarnierSillam E. Isolation and characterization of Sporobacter termitidis gen nov sp nov, from the digestive tract of the wood-feeding termite Nasutitermes lujae. Int. J. Syst. Bacteriol. 1996;46:512–518. doi: 10.1099/00207713-46-2-512. [DOI] [Google Scholar]
- 44.Shin N.R., Whon T.W., Bae J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 45.Litvak Y., Byndloss M.X., Tsolis R.M., Baumler A.J. Dysbiotic Proteobacteria expansion: A microbial signature of epithelial dysfunction. Curr. Opin. Microbiol. 2017;39:1–6. doi: 10.1016/j.mib.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 46.Johnson J.S., Spakowicz D.J., Hong B.Y., Petersen L.M., Demkowicz P., Chen L., Leopold S.R., Hanson B.M., Agresta H.O., Gerstein M., et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019;10:5029. doi: 10.1038/s41467-019-13036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study is included in the article. The raw sequences were deposited in NCBI under the Bioproject accession number PRJNA723169. Further information on data and samples is available from the corresponding author on request.