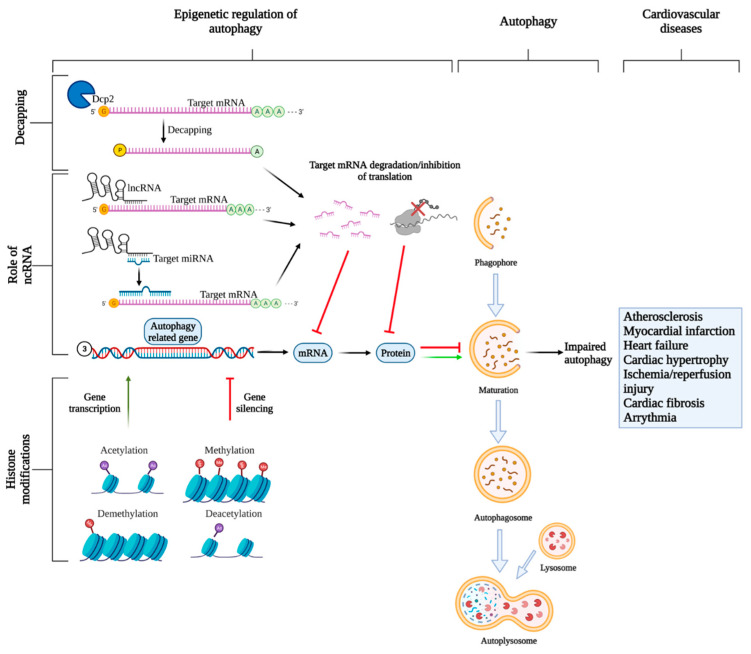
Figure 3.
Epigenetic regulation of autophagy. Three types of epigenetic regulations are discussed in this review: mRNA decapping, regulation of gene transcription and translation by ncRNAs, and histone modifications. All three processes tightly control autophagy by regulating the expression of autophagy-related genes. The decapping enzyme Dcp1 is responsible for removing the 5′cap on the target mRNAs which leads to decreased mRNA stability and consequent degradation by exonucleases. NcRNAs, such as miRNAs, can bind to target mRNAs leading to their inhibition or degradation or inhibition depending on the degree of complementarity between miRNA and mRNA. Another ncRNA, lncRNAs can control autophagy-related gene expressions either directly by interacting with target mRNAs leading to their degradation, or indirectly by acting as “miRNA” sponges to sequester miRNAs to prevent interaction with their target mRNAs allowing gene expression. Lastly, histone modifications can also influence gene expressions. Acetylation and demethylation, in most cases, lead to gene transcription, while methylation and deacetylation mostly lead to gene silencing. Reduced expression of autophagy-related genes can lead to dysregulated autophagy, which is linked to CVDs.