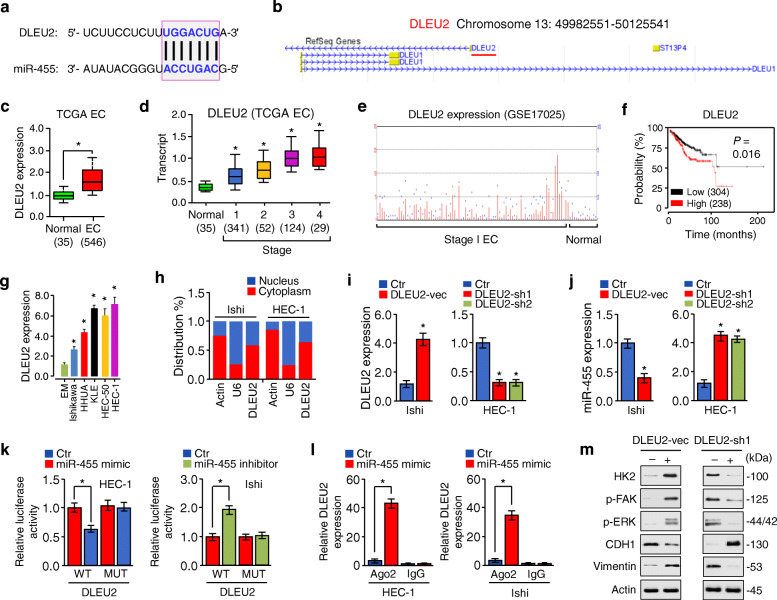
Fig. 5.
DLEU2 enhances HK2 expression via competitively binding with miR-455. a The miR-455 binding sequence in the DLEU2 sequence. b A schematic diagram shows that the DLEU2 gene is located on chromosome 13q14.2 (red bar). c Analysis of DLEU2 expression in EC and normal tissues using the TCGA data from UALCAN. d The expression of DLEU2 in normal tissues, and subgroups of patients with EC stratified based on tumor stage (UALCAN). e The NCBI Gene Expression Omnibus (GEO) dataset GSE17025 was used for profiling DLEU2 expression in stage I EC samples and normal endometrium samples. f Kaplan-Meier overall survival analysis was used to assess EC patients with high or low DLEU2 expression based on the TCGA data from KM Plotter. g qRT-PCR analysis of DLEU2 expression in the normal endometrial epithelial cell line EM and EC cell lines. h qRT-PCR analysis of the relative DLEU2 expression after nuclear and cytoplasmic RNA separation. GAPDH was used as a cytoplasmic marker, and U6 was used as a nuclear marker. i, j qRT-PCR analysis of DLEU2 (i) and miR-455 (j) expression upon overexpression or knockdown of DLEU2 in EC cells. k Luciferase reporter assay with EC cells co-transfected with a luciferase reporter plasmid containing the wild-type (WT) or mutant (MUT) DLEU2, together with miR-455 mimic, miR-455 inhibitor, or their respective controls. l RIP assays were performed in EC cells that transiently overexpressed miR-455 mimic or control mimic, followed by qRT-PCR analysis to detect DLEU2 expression associated with Ago2. m Western blotting analysis of the indicated proteins in EC cells upon overexpression or knockdown of DLEU2. Ishi: Ishikawa; vec: vector; sh: shRNA. *P < 0.05