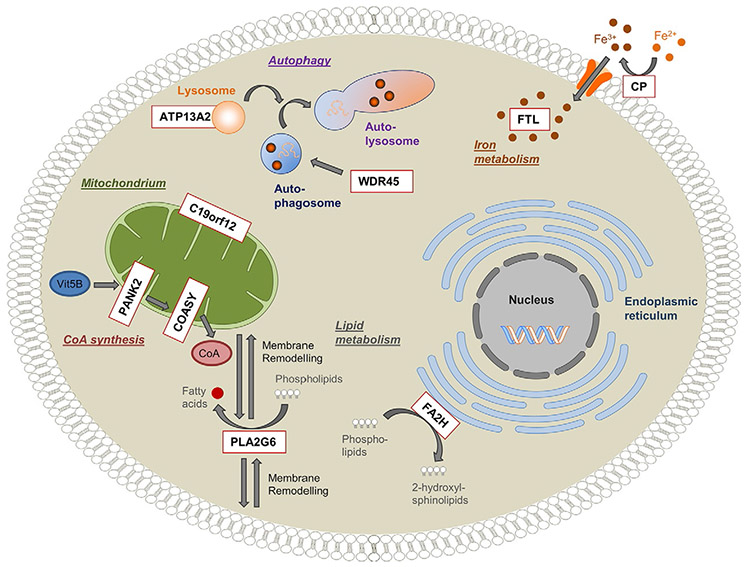
Fig. 19.2.
Overview of genes and pathways involved in neurodegeneration with brain iron accumulation (NBIA). Schematic representation of a cell indicating the pathophysiologic mechanisms postulated for NBIA. Nine reported NBIA genes (white box, red outline) and their associated cellular processes are shown. Ceruloplasmin (CP) and ferritin light polypeptide (FTL) are important regulators of cellular iron homeostasis. Phospholipase A2 (PLA2G6) and fatty acid 2-hydroxylase (FA2H) are involved in lipid metabolism and membrane remodeling. Pantothenate kinase 2 (PANK2) and CoA synthase (COASY) are key enzymes in the biosynthesis of coenzyme A (CoA) utilized in a multitude of cellular processes, including the synthesis of fatty acids. WD repeat domain 45 (WDR45) and ATPase type13A2 (ATP13A2) play a role in the degradation process of autophagy. The function of the mitochondrial membrane protein C19orf12 is still unknown, but it is postulated to be associated with fatty acid biogenesis. (Reproduced from Meyer E, Kurian MA, Hayflick SJ (2015) Neurodegeneration with brain iron accumulation: genetic diversity and pathophysiological mechanisms. Annu Rev Genomics Hum Genet 16: 8.1–8.23, with permission.)