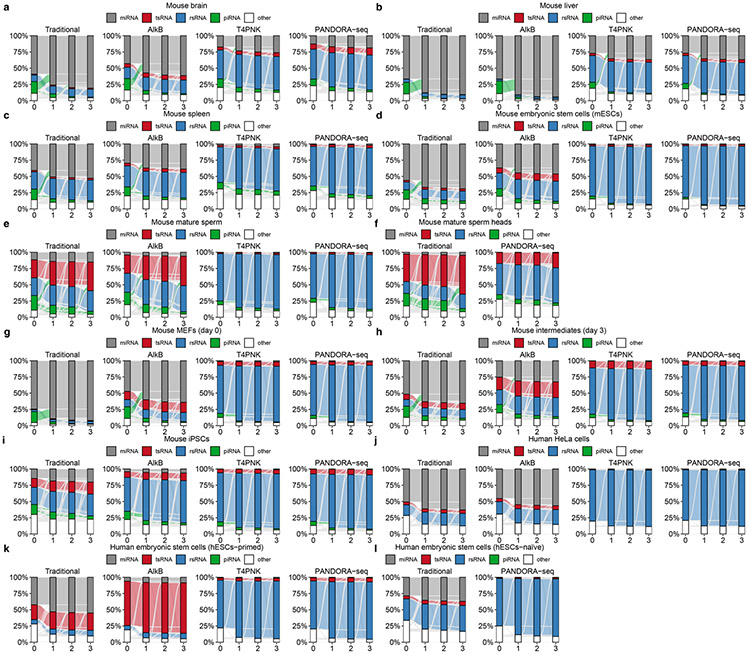
Extended Data Fig. 3 ∣. Annotation of mouse piRNA in non-germ cell tissue/cell types is not stable when 1–3 mismatches are allowed.
When 1–3 mismatches are allowed for sncRNAs matching, the piRNA annotation rate (but not other sncRNAs types) show significant decrease in mouse tissue/cell types (a) mouse brain, (b) mouse liver, (c) mouse spleen, (d) mouse embryonic stem cells, (e) mouse mature sperm, (f) mouse mature sperm heads, (g) mouse MEFs (day 0), (h) mouse intermediate cells (day 3), (i) mouse iPSCs. Very few piRNAs were annotated for human cell lines (j) human HeLa cells, (k) human hESCs-primed, and (l) human hESCs-naïve. These data suggest the annotated piRNAs in non-germ cell tissue/cell types could be due to database quality issue and their true identity awaits to be verified.