FIGURE 1.
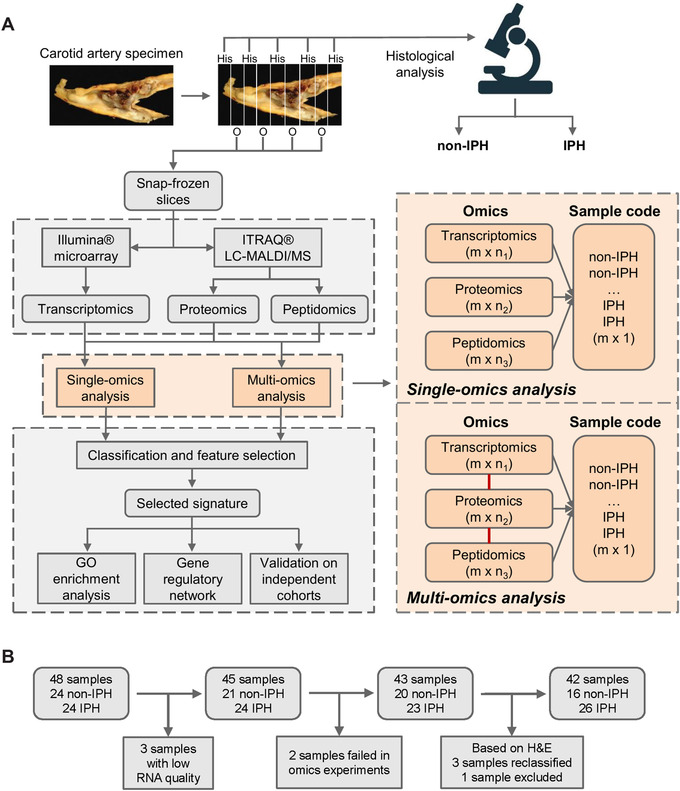
Workflow of carotid sample collection and data analysis. (A) Entire carotid endarterectomy specimen was cut in parallel 5‐mm thick slices, snap‐frozen in liquid nitrogen, and stored until use. Every second slice was sectioned. After H&E staining, sections were categorized using the criteria of Virmani et al., 41 and were further classified histologically based on the presence/absence of IPH, resulting in 16 non‐IPH and 26 IPH plaques for omics analysis. Workflow for single‐ and multiomics integrative sPLS‐DA analyses are shown in the orange dashed square. The integrative sPLS‐DA prediction model was obtained by connecting transcriptomics, proteomics, and peptidomics as an entirety; m: number of samples; n1, n2, n3: number of features. (B) Flow scheme of the MaasHPS cohort build‐up and the criteria for sample exclusion. In total, 42 samples (16 non‐IPH and 26 IPH plaques) were included in this study
