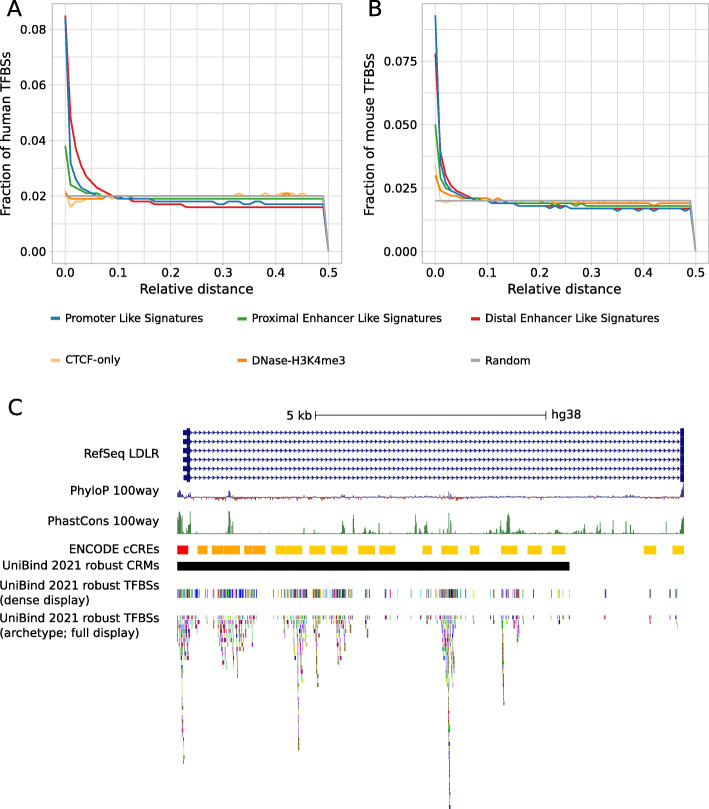
Fig. 4.
Analysis of the overlap of TFBSs with respect to active cis-regulatory regions in human and mouse. A-B Fraction of TFBSs in the UniBind robust collection (y-axis) with respect to increasing relative distances (x-axis) from ENCODE candidate cis-regulatory regions (cCREs) computed using the bedtools reldist command for human (A) and mouse (B). When two genomic tracks are not spatially related, one expects the fraction of relative distance distribution to be uniform. C Genomic tracks from the UCSC Genome Browser at the human LDLR gene locus (from start to first coding exon) providing information about PhyloP and PhastCons evolutionary conservation scores and the locations of ENCODE cCREs, UniBind CRMs, UniBind TFBSs from the robust collection (using the dense display mode to maximally condense the track) and the non-redundant collection of archetype TFBSs. Colors in the ENCODE cCREs track indicate: promoter-like signature (red), proximal enhancer-like signature (orange), and distal enhancer-like signature (yellow)