Figure 1. AAV Capsid and Genome Structure for Efficient Retrograde Access to Songbird and Mouse Projection and Dopaminergic Neurons.
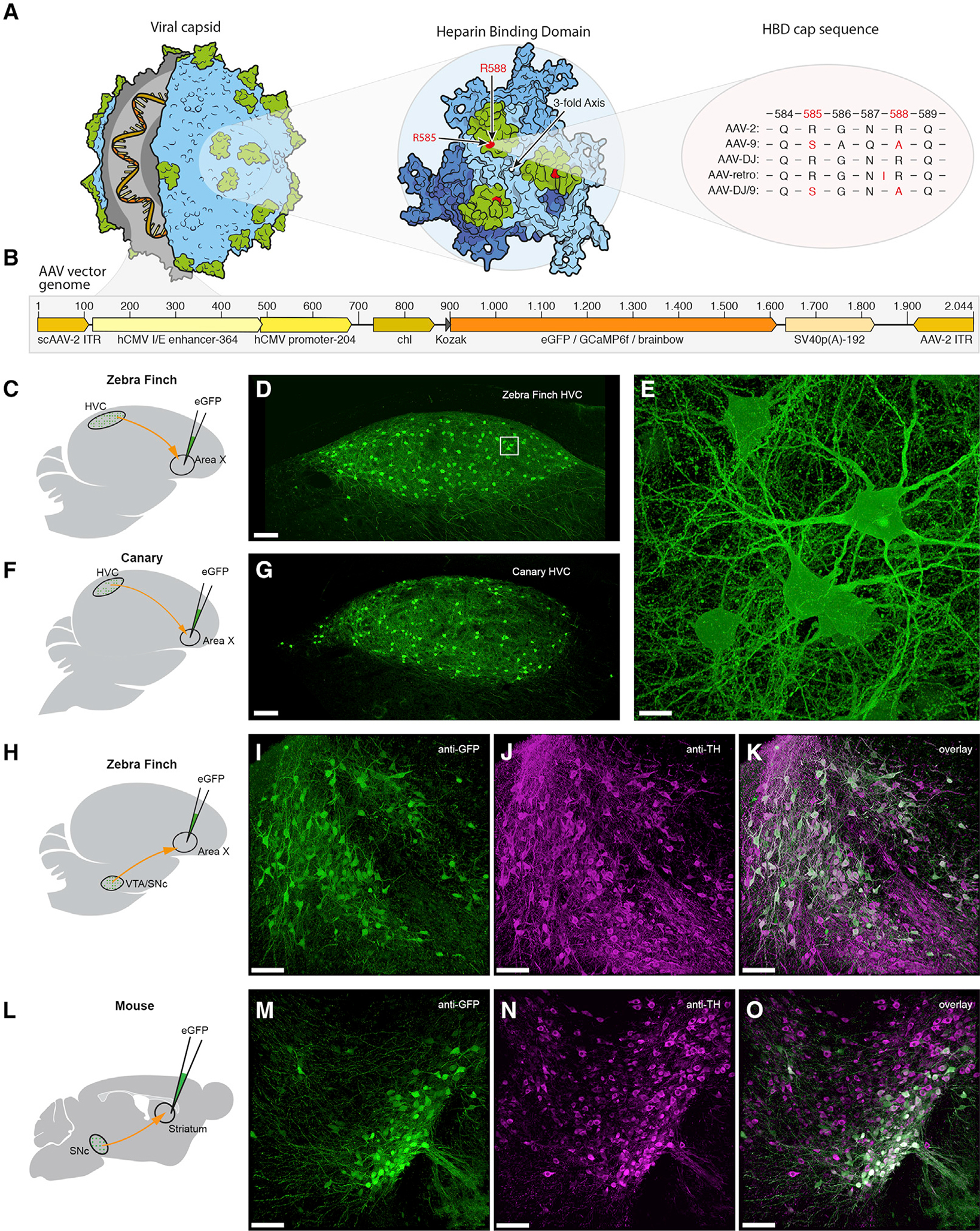
(A) Illustration of the AAV capsid, showing exposed surface proteins responsible for binding to the heparin sulfate proteoglycans (HSPG) receptor, thought to be important for cellular entry of the viral vector. Mutations of two crucial arginines in the heparin binding domain (HBD; highlighted in green) cap sequence at positions 585 and 588 (highlighted in red) reduce heparin binding affinity and potentially improve retrograde transport. Changes are shown for four serotypes that have been used for or possess potential for retrograde transduction.
(B) AAV vector genomes encoding diverse transgenes (orange region) including fluorescent proteins and the calcium indicator GCaMP6f. Numbers on top indicate the base pair length and relative position of encoded elements. Note that the actual size of the region encoding the transgene (orange) varies depending on the respective protein. Brainbow stands for three separate AAV vector genomes encoding either eGFP (enhanced green fluorescent protein), eCFP (enhanced cyan fluorescent protein), or mRuby3 (a red fluorescent protein).
(C, F, H, and L) Schematics of the virus injection sites (green) and the neuronal projections analyzed (red).
(D and G) Native fluorescence signal in retrogradely labeled HVCX neurons 7 days post-delivery of virus in zebra finch and in canary, respectively.(E, I–K, and M–O) Higher magnification z stack of the boxed region in (D). Dopaminergic projection neurons in zebra finch (I–K) and mouse (M–O) retrogradely labeled with our eGFP-construct, after immunostaining for anti-GFP (I and M) and anti-tyrosine hydroxylase (J and N). Scale bars: 100 μm in (D), (G), (I)–(K), and (M)–(O) and 10 μm in (E).
