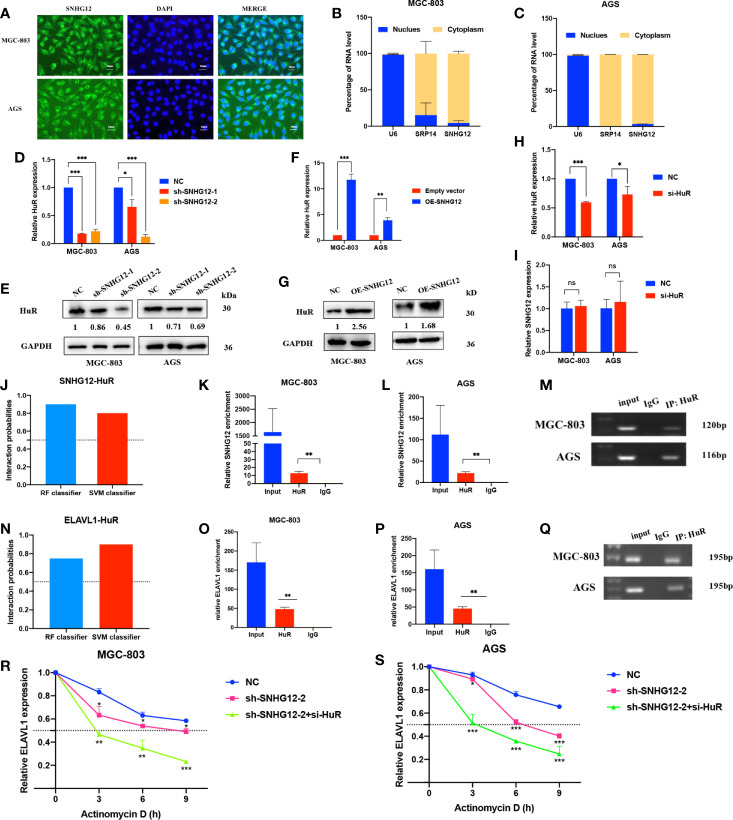
Figure 3.
SNHG12 binds to HuR and stabilizes ELAVL1 mRNA. (A–C) FISH assays and cytoplasmic and nuclear RNA purification assays indicate that SNHG12 is mainly located in the GC cell cytoplasm. (D–G) qRT-PCR and WB assays showing the expression of HuR at the RNA and protein levels upon SNHG12 knockdown and overexpression. Numbers show the quantification of the relative protein amount. (H, I) qRT-PCR assays showing the efficiency of HuR knockdown and its effect on the regulation of SNHG12. (J) Prediction of the interaction probabilities of SNHG12 and HuR by bioinformatics (http://pridb.gdcb.iastate.edu/RPISeq/). Predictions with probabilities >0.5 were considered as “positive”, indicating that the RNA more likely to interact with the protein than not to interact. (K–M) RIP assays showing that SNHG12 binds to HuR. (N) Prediction of the interaction probabilities of ELAVL1 and HuR by bioinformatics (http://pridb.gdcb.iastate.edu/RPISeq/). (O–Q) RIP assays showing that ELAVL1 binds to HuR. (R, S) RNA stability assays were conducted using actinomycin D to disrupt RNA synthesis in MGC-803 and AGS cells, and the degradation rates of ELAVL1 mRNAs were tested every 3 h. Significant results are presented as ns P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001. Magnification ×400; scale bar, 20 μm.