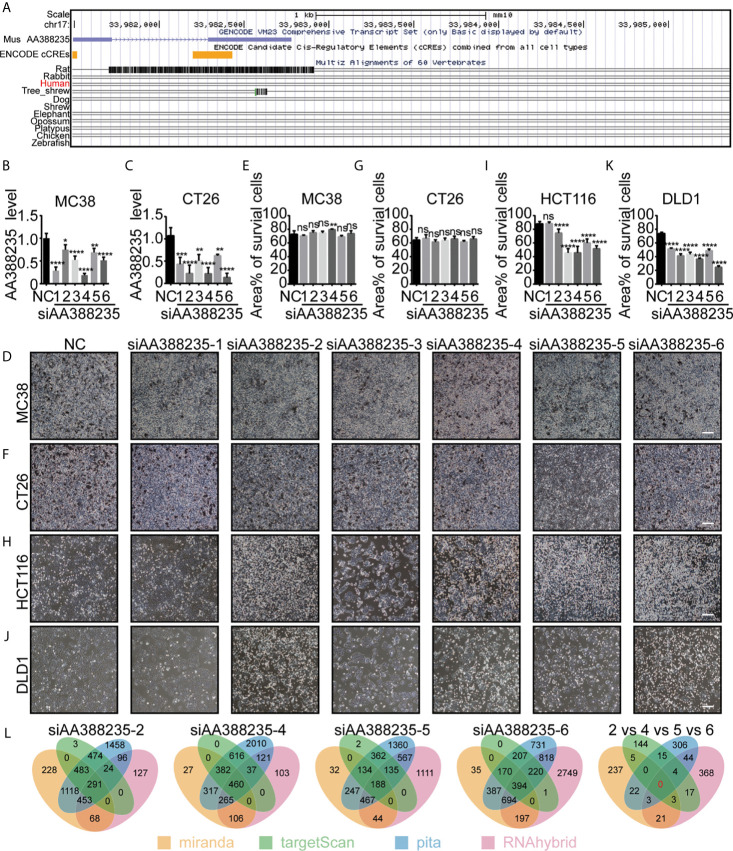
Figure 1.
siRNAs targeting mouse-specific lncRNA AA388235 induce human colorectal tumor cells death. (A) Graphical views showing multiple-species conversation comparisons with mouse LncRNA AA388235 using UCSC genome browser. The conservation scores were indicated by the gray peaks. (B, C) ”NC” was a random arrangement and meaningless 19 bp siRNA, which was desired by RiboBio to act as the negative control.”siAA388235-1/2/3/4/5/6”meant six 19 bp siRNAs targeting six different regions of mouse LncRNA AA388235. The AA388235 level in MC38 and CT26 cells transfected with siRNAs as indicated. P value was calculated by one-way ANOVA. (D–G) MC38 and CT26 were seeded and treated as indicated. After 48 h, the static bright-field cell images were captured (D, F). Then the dead cells were washed off with PBS, and the living cells were captured to measure the area percentage of survival cells using Image J software from five independent perspectives (E, G). Scale bar, 200μm. P value was calculated by one-way ANOVA. (H–K) HCT116 and DLD1 were seeded and treated as indicated. After 48 h, the static bright-field cell images were captured (H, J). Then the dead cells were washed off with PBS, and the living cells were captured to measure the area percentage of survival cells using Image J software from five independent perspectives (I, K). Scale bar, 200μm. P value was calculated by one-way ANOVA. (L) Prediction of off-target genes of siAA388235 using database: miranda, targetScan, pita, and RNAhybrid. “2 vs 4 vs 5 vs 6” meant the prediction of common off-target genes between the siAA388235-2,4,5, and 6. There were no common off-target genes between the siAA388235-2,4,5, and 6 in four databases and ten off-target genes in random three databases including AHRR, C3ORF14, SLC15A2, COQ8A, FAM46C, GOLGA8H, TNRC6C, TLK2,PAGR1, PGS1. Mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, “ns” indicates no significance.