Fig. 6. Experimental investigation of alleles with perturbed physical protein-protein interactions.
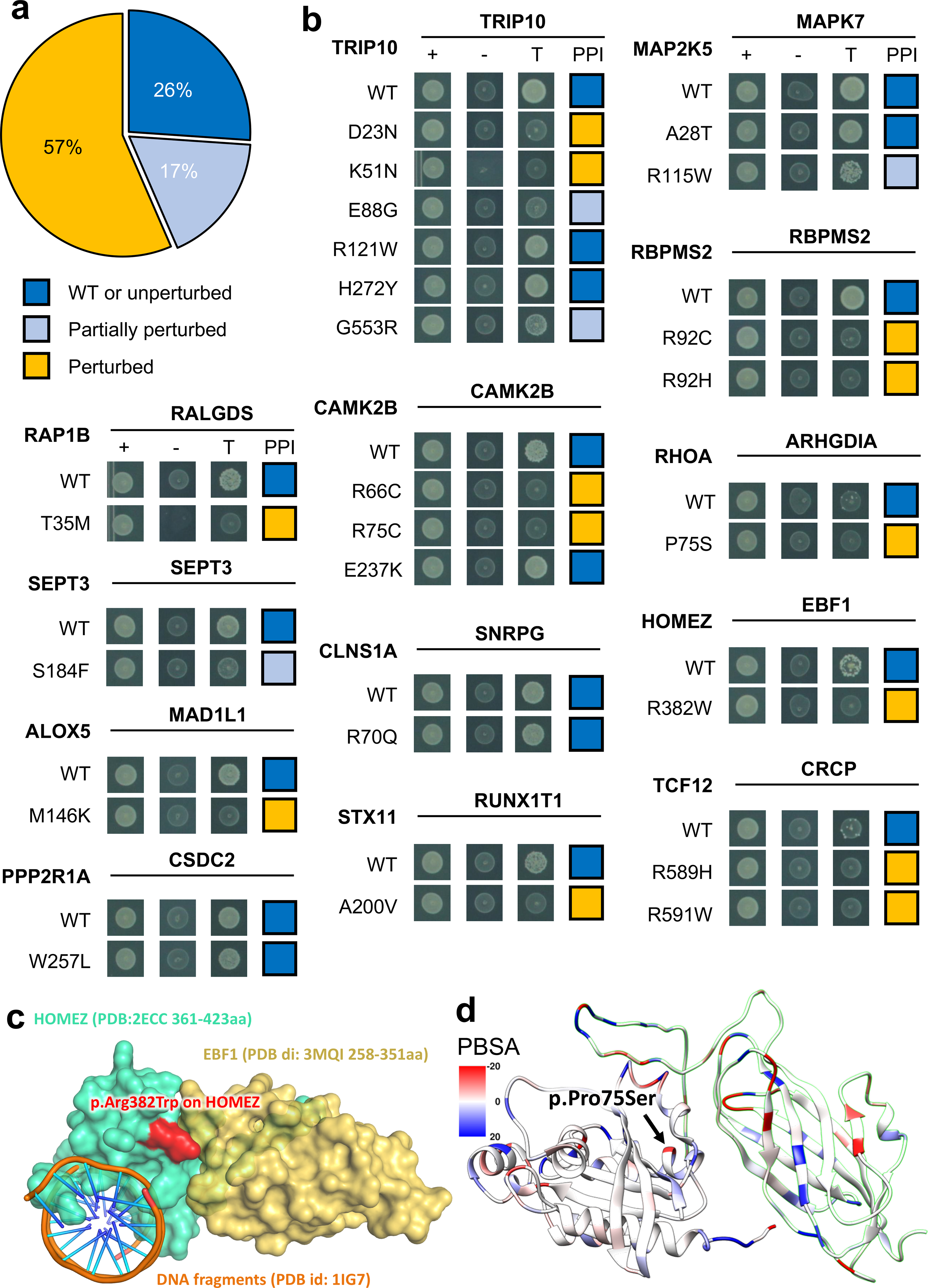
(a) Distribution of three types of mutational consequences on PPIs, unperturbed, partially perturbed, and perturbed. (b) Y2H readouts of oncoPPIs with and without mutations. “+” represents selection for existence of AD and DB plasmids that carry ORFs for PPI testing, “-” represents selection for auto-activators, “T” represents selection for PPIs. Growth indicates interaction, no growth suggests no interaction (see Methods and Supplementary Table 3). Growth indicates interaction, no growth suggests no interaction (see Methods and Supplementary Table 3). (c) HOMEZ-EBF1 complex model and the location of the interface mutation, p.Arg382Trp on HOMEZ. The complex model was built by Zdock protein docking simulation (see Supplementary Figure 18). (d) Distribution of calculated binding affinity (PBSA) of RHOA-ARHGDIA complex (PDB id: 1CC0) directed by p.Pro75Ser mutation on RHOA. Color bar indicates binding affinity (see Methods) from high (blue) to low (red). WT: wild-type.
