Fig. 1.
Study design and multiplatform metabolomics data generation.
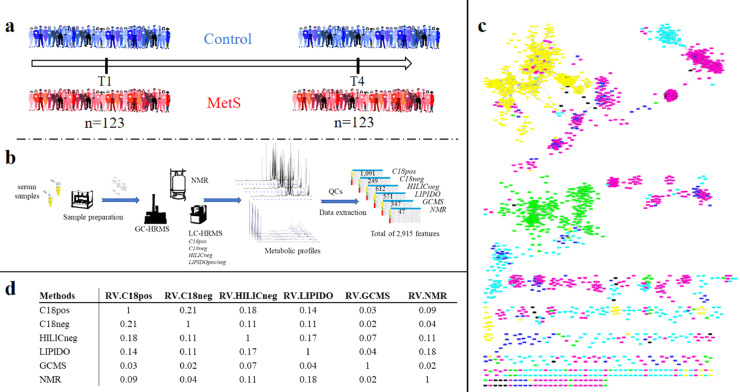
a: Experimental design of the MetS case/control study, involving the follow-up of 123 stable subjects over 3 years.
b: Analytical workflow based on seven complementary untargeted metabolomics methods using 3 different analytical platforms (LC- and GC-HRMS, NMR) for the serum analysis of the 123 subjects collected at two time points. Data analysis resulted in 2915 metabolite and lipid related features, detected across at least 20% of the subjects over time.
c: The similarity of method blocks evaluated using RV coefficients (a matrix correlation coefficient (see method section)) after Multiple Factorial Analysis. d: Spearman correlation network between the 2915 metabolite and lipid features from the six metabolomic/lipidomic datasets (significant correlation >0.7). Nodes correspond to features obtained using the different analytical platform (Pink: C18pos; Dark Blue: C18neg; Green: GCMS; Blue: HILICneg; Yellow: Lipidomics; Black: NMR). Two nodes are connected by an edge if correlation is significant between the corresponding features (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).