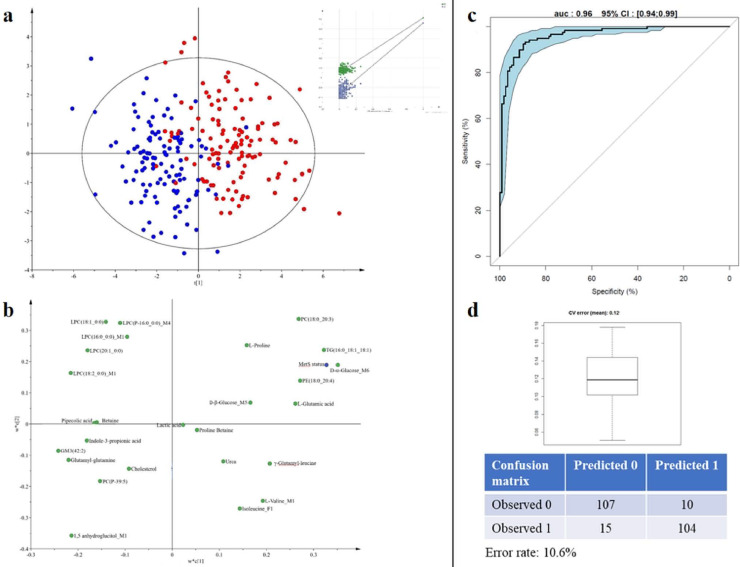
Fig. 6.
Reduced MetS signature.
a: Score plot of the Partial Least squares Discriminant Analysis performed on the most robust unique annotated compounds for all subjects at both times (R2Y=0.58 and Q2=0.56) with the graphical result of the 200-permutation evaluation.
b: Loading plot associated with the score plot of the PLS-DA. This plot corresponds to the projection of each variable on the latent components of the PLS-DA model. The farthest a variable is from the plot origin, the highest importance it has to the discrimination. As the twin of the score plot, the loading plot enables to grasp similarity between variables in the contribution to discrimination (proximity on the plot) as well as to assess which group of individuals is more likely to have a high intensity value of the metabolite. Status: MetS status; Metabolite_M1: detected in C18pos; Metabolite_M2: detected in C18neg; Metabolite_M3: HILICneg; Metabolite_M4: detected in LIPIDO; Metabolite_M5: detected in GCMS; Metabolite_M6 detected in NMR; Metabolite_F: variable related to a metabolite fragment.
c: ROC curve of the PLS-DA model (11% adjusted misclassification rate, AUC=0.96, CI:[0.94–0.99]). d: box-plot of the error rate corresponding to the 200 cross-validated models (12% error rate, with adjusted confusion matrix). Graphical representation using the SIMCA software – Umetrics AB, 2015.