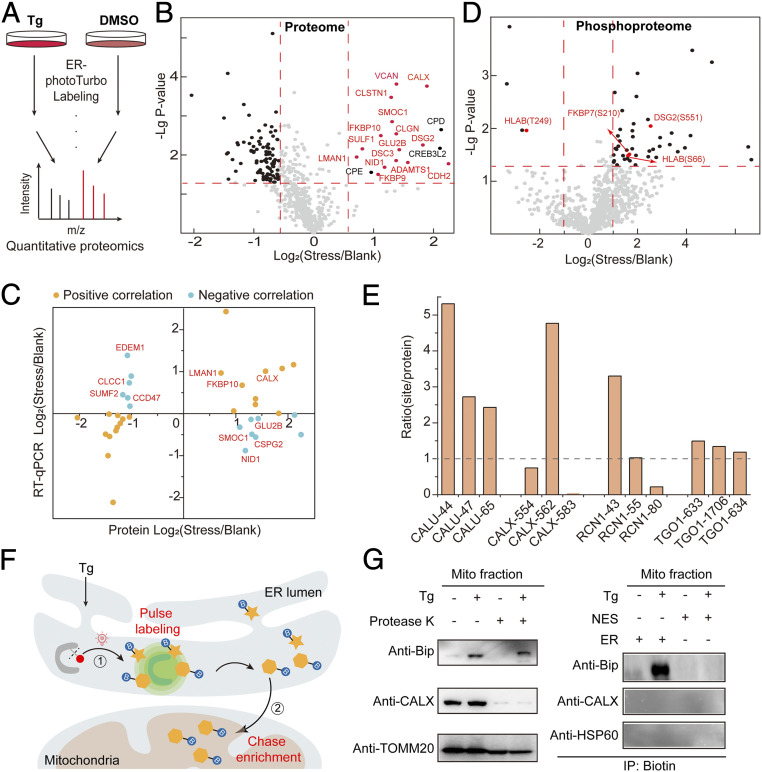
Fig. 4.
SubMAPP for dynamic subcellular proteomics and phosphoproteomics. (A) Experimental scheme for ER-photoTurbo labeling in the presence and absence of ER stress inducer, Tg. (B) Volcano plot showing significant change of secretory proteins in ER lumen under 500 nM Tg treatment. Calcium binding proteins are highlighted in red. (C) The transcription and translation correlation analysis on proteins under ER stress. (D) Volcano plot showing the change of phosphorylated peptides in ER lumen under ER stress. (E) The correction of phosphorylation level of partial phosphorylated proteins, which coincide with ER proteome under ER stress. (F) Schematic description of the photo-controlled Turbo-enabled “pulse-chase” strategy for monitoring or identification of ER-to-mitochondria translocated proteins. In brief, 1) ER proteins were first “pulse” labeled by photo-activation of Turbo upon ER stress, and 2) the mitochondria fraction was then isolated to “chase” enrich the biotinylated proteins in mitochondria. (G) Immunoprecipitation for detecting the ER-to-mitochondria translocated Bip protein under ER stress. Mitochondria fraction was isolated by mitochondria isolation kit (Abbike KTP4003) and treated with protease K before being subject to immunoblotting analysis. CALX was used as the ER marker, while TOMM20 and HSP60 were used as the mitochondrial markers.