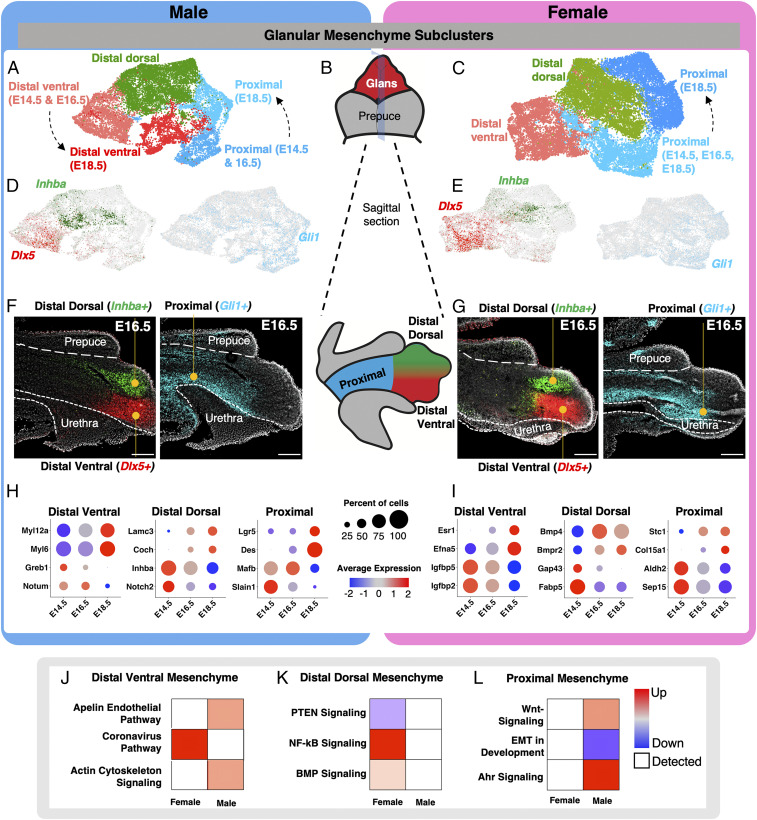
Fig. 3.
Identification of unique subpopulations in the glanular mesenchyme. (A and C) Reclustering of the glanular mesenchymal cells in male and female from Fig. 2 with higher resolution. The different colored cells represent distinct cell clusters identified with unbiased clustering. (B) Whole mount and sagittal-section representation of external genitalia that highlight the glans in red (whole mount), external prepuce in gray (whole mount), proximal glans in blue (sagittal), distal dorsal glans in green (sagittal), and distal ventral glans in red (sagittal). (D and E) UMAPs of glanular mesenchyme subclusters in A and C, respectively, that are positive for Inhba (green), Dlx5 (red), and Gli1 (light blue). (F and G) In situ hybridization for Inhba (green), Dlx5 (red), and Gli1 (light blue) on sagittal sections of external genitalia. The external prepuce and urethra portions of the external genitalia are outlined with dotted lines and with the glans in the center. (H and I) Dot plots of marker genes that are enriched in different cell populations in the male and female genitalia from A and C, respectively. The dot size represents the percentage of cells that express the gene, and the color represents the intensity of expression (red = high and blue = low). (J–L) Pathway analysis of genes that were identified in H and I. The pathways were selected based on whether they were unique to male or female development. Red = up-regulated, blue = down-regulation, white = detected, and circle size = P value of the pathway. (Scale bars, 100 µm.)