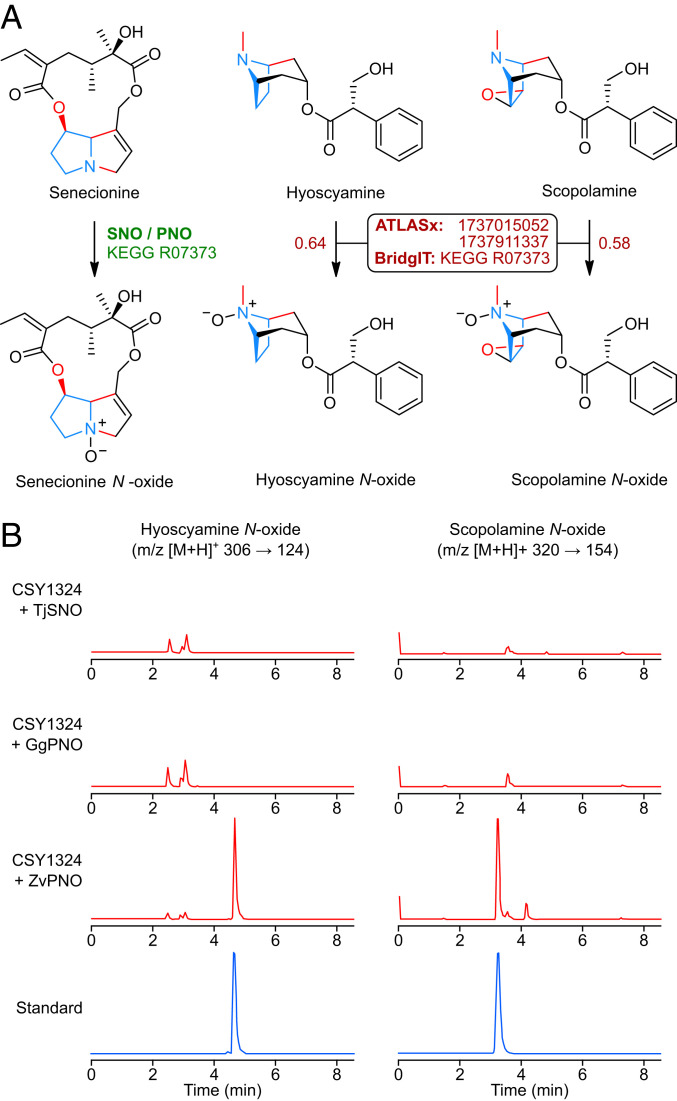
Fig. 7.
In silico enzyme prediction using BridgIT enables de novo biosynthesis of TA N-oxides in engineered yeast. (A) Comparison of native reaction (Left) and target reactions (Center, Right) for SNO and PNO, the top-scoring enzymes predicted by BridgIT to N-oxygenate TAs. The KEGG reaction ID for native SNO/PNO transformation is shown in green; ATLASx reaction IDs and the BridgIT closest KEGG reaction match for target transformations are shown in dark red. Values beside arrows for target reactions indicate BridgIT scores. Conserved pyrrolidine core and shared functional group connectivity in substrates and products are indicated in blue and red, respectively. (B) LC–MS/MS multiple reaction monitoring chromatograms for analysis of de novo TA N-oxide production by CSY1324 expressing SNO/PNOs, or authentic chemical standards. SNO/PNO candidates from T. jacobaeae (TjSNO), G. geneura (GgPNO), and Z. variegatus (ZvPNO) were expressed from low-copy plasmids in CSY1324, and metabolites in supernatant were analyzed after 96-h growth in selective media buffered to pH 5.8 with 0.1 M potassium phosphate (for hyoscyamine N-oxide) or unbuffered (for scopolamine N-oxide). Chromatograms are representative of n = 3 biologically independent samples.