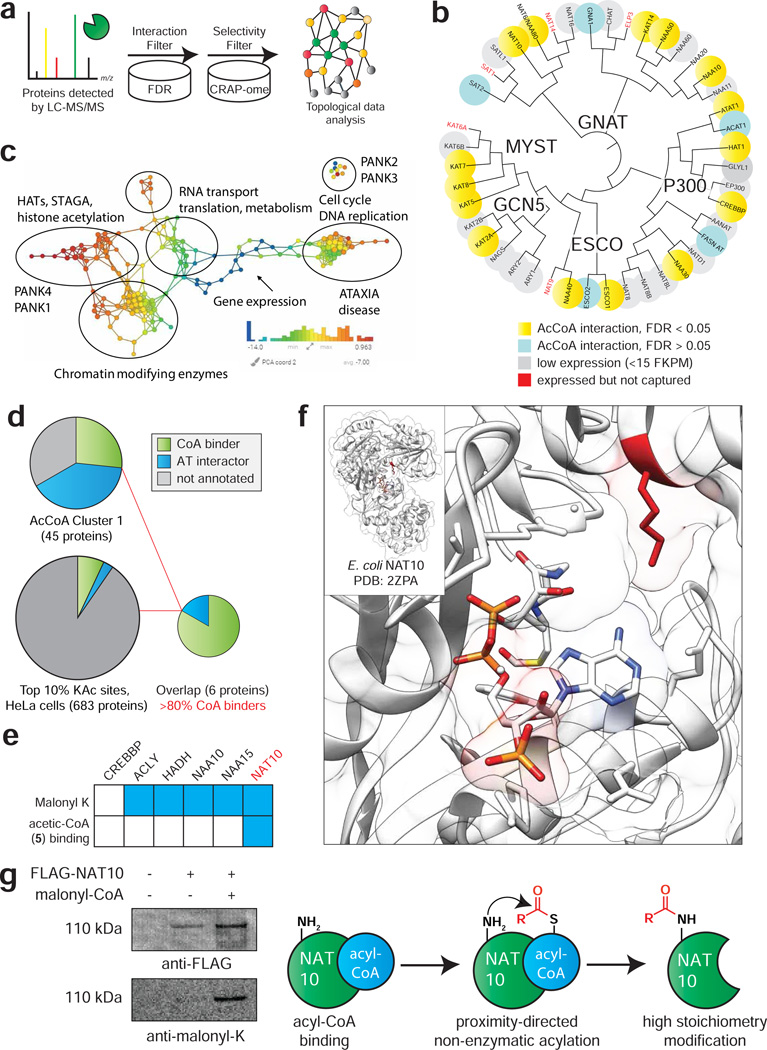
Figure 5.
Applying CATNIP to assess the annotation of the CoA-binding proteome. (a) Schematic for differentiating CATNIP-enriched proteins from background based on significant competition and absence from common contaminant databases. (b) Proteins from diverse AT families display statistically significant CoA/acyl-CoA competition. (c) Topological network analysis of proteins exhibiting significant interaction with ≥ 3 CoA metabolites. Protein nodes are colored based on the metric PCA2. Color bar: red = high values; blue = low values. Node size is proportional to the number of proteins in the node. (d) Combining CATNIP competition (QPROT Log2FC ≤ −2 and FDRdown<0.05) and acetylation stoichiometry filters greatly enriches CoA-binders and AT-interactors relative to either measure alone. (e) Comparing annotated sites of lysine malonylation (top) and competition by malonyl-CoA mimic 5 (bottom) for CoA-binders detected by de novo CATNIP analysis. (f) A conserved site of lysine malonylation lies in close proximity to the acetyl-CoA binding site of bacterial NAT10. (g) Overexpressed FLAG-NAT10 is malonylated in the presence of malonyl-CoA, consistent with a non-enzymatic acylation mechanism.