Figure 1.
The core microbiome
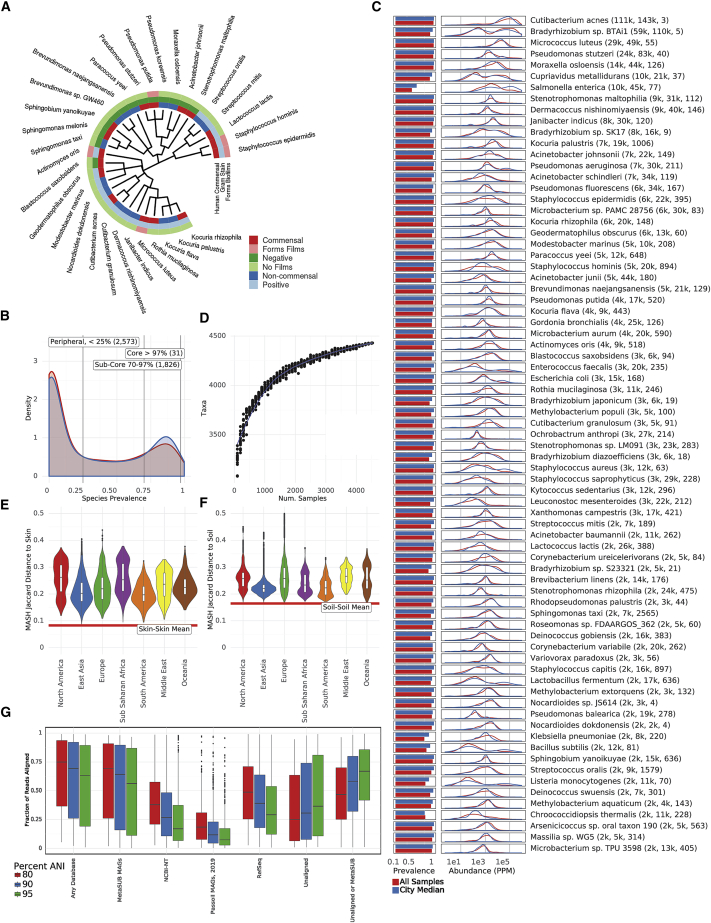
(A) Taxonomic tree showing 31 core taxa, annotated according to gram stain, ability to form biofilms, and whether the bacteria is a human commensal species.
(B) Distribution of species prevalence from all samples and normalized by cities. Vertical lines show defined group cutoffs.
(C) Prevalence and distribution of relative abundances of the 75 most abundant taxa. Mean relative abundance, standard deviation, and kurtosis of the abundance distribution are shown.
(D) Rarefaction analysis showing the number of species detected in randomly chosen sets of samples.
(E) MASH (k-mer-based) similarity between MetaSUB samples and HMP skin microbiome samples by continent.
(F) MASH (k-mer based) similarity between MetaSUB samples and soil microbiome samples by continent.
(G) Fraction of reads aligned (via BLAST) to different databases at different average nucleotide identities.
See also Figure S1.