Figure 3.
Microbial signatures
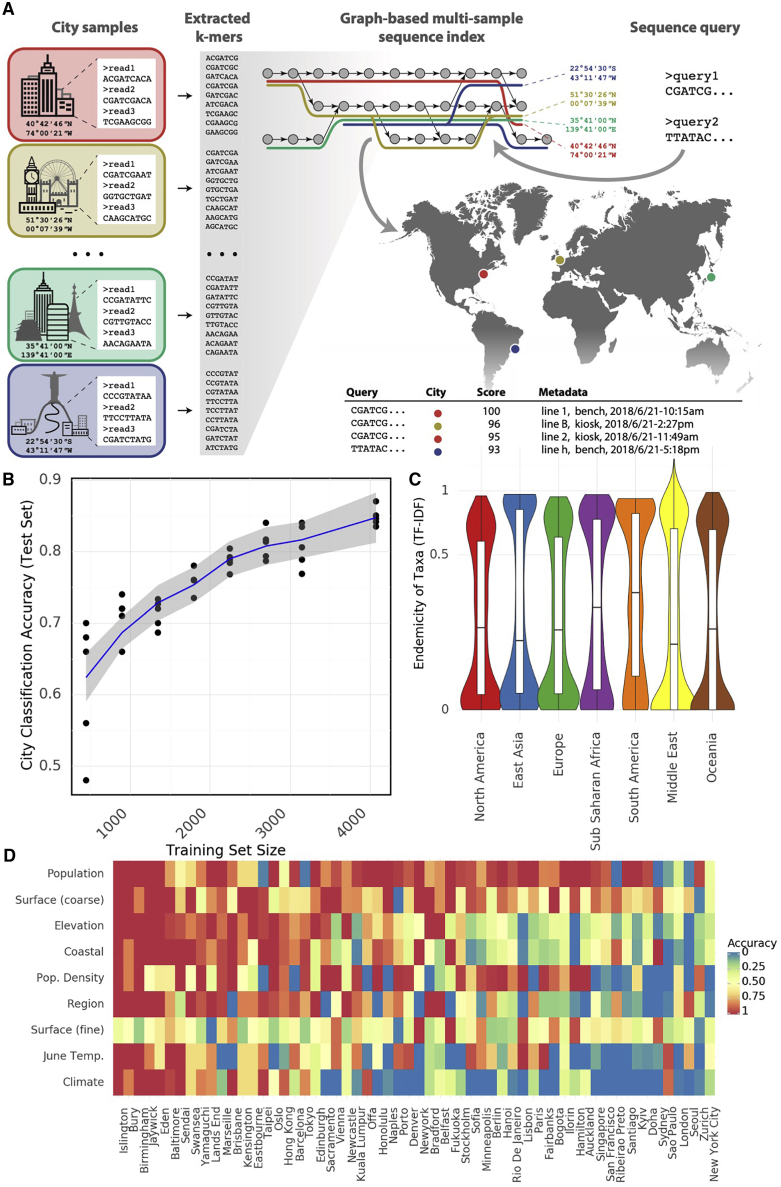
(A) Schematic of GeoDNA representation generation. Raw sequences of individual samples for all cities are transformed into lists of unique k-mers (left). After filtration, the k-mers are assembled into a graph index database. Each k-mer is then associated with its respective city label and other informative metadata, such as geo-location and sampling information (top middle). Arbitrary input sequences (top right) can then be efficiently queried against the index, returning a ranked list of matching paths in the graph together with metadata and a score indicating the percentage of k-mer identity (bottom right). The geo-information of each sample is used to highlight the locations of samples that contain sequences identical or close to the queried sequence (middle right).
(B) Classification accuracy of a random forest model for assigning city labels to samples as a function of the size of the training set.
(C) Distribution of endemicity scores (term frequency inverse document frequency) for taxa in each region.
(D) Prediction accuracy of a random forest model for a given feature (rows) in samples from a city (columns) that were not present in the training set. Rows and columns are sorted by average accuracy. Continuous features (e.g., population) were discretized.
See also Figure S4.